Abstract
Several studies suggest membrane trafficking events are mediated by integral, membrane proteins from both transport-vesicle and target membranes, called v- and t-SNAREs (SNAp REceptors), respectively. Previous experiments using antibodies to synaptobrevin/vesicle associated membrane protein (VAMP) 1, 2, or rat cellubrevin failed to detect these v-SNAREs in human platelets, although membrane proteins from these cells could support 20S complex formation. To identify v-SNAREs in platelets, we used a polymerase chain reaction (PCR) approach with degenerate primers to amplify potential VAMP-like v-SNAREs. A cDNA encoding a novel v-SNARE was isolated from a human megakaryocyte cDNA library. Termed human cellubrevin (Hceb), this protein has greater than 93% identity with human VAMP 1, 2, and rat cellubrevin over the conserved core region, but has a unique N–terminal domain. Northern blot analysis showed that the 2.5-kB mRNA encoding Hceb is expressed in every human tissue tested. Hceb from detergent-solubilized platelet membranes, participated in -SNAP–dependent 20S complex formation and adenosine triphosphate (ATP)-dependent disassembly, showing that Hceb can act as a v-SNARE in platelets. Immunofluorescence microscopy, using an anti-Hceb antibody showed a punctate, intracellular staining pattern in platelets, megakaryocytes, and HEK-293 cells. This same pattern was observed in surface-activated platelets even though all dense core and most -granule contents had been released. These data suggest that Hceb may reside on a platelet organelle that is not primarily involved in the exocytic pathway.
ONCE ACTIVATED, PLATELETS secrete components important for clot formation and wound repair. Of special importance to their hemostatic function are their granules. It is the alpha and dense core granules that contain the components necessary for hemostasis. Binding to blood vessel lesions activates platelets causing the granules to concentrate to the center of the cell. One theory proposes that the concentrated granules fuse with specialized regions of the plasma membrane called the open canalicular system (OCS),1 thereby releasing the granule contents into channels that lead to the extracellular space. An alternative model proposes that the OCS is first extruded into the plasma membrane, then the granules concentrate to the center of the cell where they undergo compound fusion and subsequently fuse with the plasma membrane.2 Regardless of the mechanism, these fusion events result in the release of the contents of the alpha granules and dense core granules into the extracellular space.3
Platelets also appear to undergo constitutive and regulated endocytosis. Clathrin-coated vesicles budding from the OCS have been observed by electron microscopy4 and electron-dense, fluid-phase markers can be detected in platelet granules.4To date, the most studied example of endocytic traffic involves the receptor-mediated internalization of fibrinogen via the fibrinogen receptor (gpIIβ/IIIα). Because megakaryocytes and platelets do not synthesize fibrinogen,5,6 all of the fibrinogen in the alpha granules must be endocytosed from the plasma.7Kinetic studies of fibrinogen uptake into megakaryocytes, established the multivesicular bodies (MVBs) as endocytic, precursors of the alpha granules.8 Platelets also endocytose fibrinogen from the plasma.7,9 Endocytosis in resting platelets occurs constitutively at a slow rate,9 but the rate is increased when platelets are partially stimulated with agonists such as adenine diphosphate (ADP) or low concentrations of thrombin.10Although platelets contain MVBs, the precursor relationship to alpha granules has not been established.8
Studies of membrane trafficking in mammalian systems and in yeast have suggested that docking and fusion of transport vesicles with target membranes are mediated by soluble proteins (ie, N-ethylmaleimide sensitive factor (NSF), α and γ-soluble NSF attachment proteins (SNAPs)) and by integral membrane proteins, known as SNAp REceptors or SNAREs.11-13 The synaptobrevin/vesicle associated membrane protein (VAMP) family of SNARE proteins reside on the vesicle and are therefore termed, v-SNAREs. The v-SNAREs, VAMP 1 and 2 and rat cellubrevin, are small molecular weight (≈ 18 kD), type II integral membrane proteins with a unique N-terminus and two highly conserved coiled-coil domains, which are important for targeting VAMPs to vesicles14 and for binding to t-SNAREs.15,16The syntaxins and synaptosomal associated protein (SNAP)-23/25 family members are thought to form a heterodimer on the target membrane and are therefore called t-SNAREs.17 Since the discovery of the SNARE proteins in neurons, homologues have been found in a number of different tissues, and importantly, on different membranes within a cell.18 The original SNARE hypothesis proposed that for each vesicular trafficking event there will be a specific v-SNARE that binds to its cognate t-SNARE heterodimer.12,19 Once the v- and t-SNAREs bind, they form a 7S complex, which can interact with α-SNAP and NSF. This series of binding interactions results in the formation of the 20S complex.20 Adenosine triphosphate (ATP) hydrolysis by NSF mediates 20S complex disassembly and is a required step for membrane trafficking events in vitro.21,22 Recent studies have indicated that this complex disassembly represents a priming step that is integral to the activation of SNAREs for membrane fusion.23-26
To understand more about the membrane trafficking events in platelets, we have continued to identify proteins that could mediate platelet exocytosis or endocytosis. In the present study, we report the identification of a novel VAMP/synaptobrevin family member. Biochemical studies show that this v-SNARE is present in platelet membranes and can participate in 20S complex assembly and ATP-dependent disassembly. Originally cloned from a human megakaryocyte cDNA library, we find by Northern blotting analysis that this v-SNARE is expressed in many human tissues. Due to its high homology to other known VAMPs, its broad tissue distribution, and its subcellular localization, this protein appears to be the human equivalent of the rodent cellubrevin. In platelets, our studies show that Hceb resides on a compartment that is not mobilized to the plasma membrane on calcium or thrombin stimulation. The functional significance of a platelet cellubrevin will be discussed.
MATERIALS AND METHODS
Antibodies, cell lines, and reagents.
Oligonucleotide polymerase chain reaction (PCR) primers were from GibcoBRL (Grand Island, NY). The cDNA library was constructed with mRNA from CMK cells that were induced to differentiate with phorbol 12-myristate 13-acetate for 4 hours and was generously provided by Dr Takeyuki Sato through Mochida Pharmaceutical Co, LTD, Tokyo, Japan.27 The multiple tissue Northern blot was from Clonetech (Palo Alto, CA). Outdated platelets were procured as units from the Central Kentucky Blood Center (Lexington, KY). HEK-293 cells were a generous gift of Dr L.B. Hersh (University of Kentucky, Lexington, KY). HepG2 cells were a generous gift of Dr D.J. Noonan (University of Kentucky, Lexington, KY). Anti-VAMP 1 antibody (Ab)28 and anti-VAMP 2 Ab (C1 69.1)29 were kindly provided by Dr Reinhard Jahn (Yale University, New Haven, CT). Antirat cellubrevin Ab (MC16) was kindly provided by Dr P. DeCamilli (Yale University, New Haven, CT).28 Antitransferrin receptor Ab (MAB1451) was purchased from Chemicon International, Inc (Temecula, CA). Anti-gpIIβ/IIIα Ab (N77140M) was purchased from Biodesign (Kennebunkport, ME). Antiserotonin Ab (18-0077) was purchased from Zymed (San Francisco, CA). Anti-von Willibrand Factor Ab was purchased from Sigma (St Louis, MO).
Cloning of cDNA encoding human cellubrevin.
The plasmid-based, CMK cDNA library was divided into 15 pools containing ≈ 70,000 clones each (1.05 × 106 clones total). Each pool was analyzed by PCR using the following degenerate primers: 5′-GGAATTCC CA(AG) CA(AG) AC(TCA) CA(AG) GC(TCA) CA(AG) G-3′ and 5′-CGGGATC CAT (TC)TT (AG)CA (AG)TT (TC)TT CCA C-3′, based on conserved regions of human, bovine, andDrosophila VAMP proteins (see Fig 1B).30 PCR was run for 30 cycles using a 45°C annealing step and 1 μg of template DNA from each of the library pools. One pool gave a positive PCR product of the predicted size and on DNA sequencing proved to be homologous to known VAMPs. Colonies from the positive pool were then screened by filter hybridization with standard procedures using a [32P]-cytidine triphosphate (CTP; NEN, Boston, MA) labeled riboprobe (Promega, Madison, WI) made from the initial PCR product.
Production and purification of antihuman cellubrevin antibody.
The N-terminal human cellubrevin (Hceb) peptide, MSTGPTAATGSN-(C), was synthesized at the Macromolecular Structural Analysis Facility (Lexington, KY) and coupled to Imject Activated BSA Super Carrier (Pierce, Rockford, IL). Anti-Hceb Ab was produced by injecting the conjugate into New Zealand White rabbits at the Division of Laboratory Animal Resources facility (Lexington, KY). To affinity purify the Hceb Ab, the cDNA encoding the cytosolic domain of Hceb (a.a. 1-75) was subcloned into the pGEX-KG vector (Pharmacia, Piscataway, NJ). The Hceb-glutathione-S-transferase (GST) fusion protein was expressed in Escherichia coli (E. coli), bound to glutathione agarose beads (Sigma), and cross-linked to the beads with Bis(Sulphosuccinimidyl)suberate (2 μg/mL) (Pierce). The sera from the immunized rabbit were separated by a 50% ammonium sulfate precipitation, brought up in 50% the initial volume in phosphate-buffered saline (PBS), and dialyzed overnight against PBS at 4°C. The dialyzate was passed over the GST-Hceb fusion protein column and the bound antibody was eluted with 100 mmol/L glycine pH 2.5. The purified antibody was further incubated with GST-bound glutathione agarose beads to adsorb any contaminating anti-GST reactivity. To test the specificity of the anti-Hceb antibody, the cDNAs encoding human VAMP 1 and 2 were amplified from a human QUICK Clone spleen cDNA library (Clonetech, Palo Alto, CA) using PCR primers made to their N and C terminus (Accession No. J05611). The correct products encoding VAMP 1, 2 and Hceb were subcloned into a pQE-9 vector (Qiagen, Valencia, CA) and expressed in E. coli.
SNARE activity assay.
Human platelet membrane extract (HPE) was prepared as described by Lemons et al.31 Briefly, human platelet membranes are salt washed to remove all of the peripheral proteins and then detergent solubilized to produce a HPE. 20S Complexes were formed by incubating HPE (5 mg) with recombinant α-SNAP (100 μg), recombinant γ-SNAP (10 μg), and recombinant C-terminal-myc-tagged NSF (50 μg) in binding buffer (20 mmol/L Hepes/KOH pH 7.0, 100 mmol/L KCl, 1% polyethyleneglycol, 1% glycerol, 1% Triton-X 100, 1 mmol/L dithiothreitol [DTT]) with 1 mmol/L ATPγS and 1 mmol/L EDTA for 30 minutes on ice. In these reactions, membrane extract must constitute 50% or less of the total reaction volume to achieve noninhibitory protein/detergent ratios. NSFmyc containing complexes were then recovered by adding a saturating amount of anti-myc antibody (9E10)32 coupled to Protein G Superose beads (Pharmacia), and incubated for an additional 2 hours at 4°C. The beads were washed four times at room temperature (RT) with 0.5 mL of binding buffer with 1 mmol/L ATPγS and 4 mmol/L MgCl2 (wash, see Fig 5). The 20S complexes bound to beads were then incubated for 1 hour at 4°C with 1 mL of binding buffer with 5 mmol/L ATP and 4 mmol/L MgCl2 (release, see Fig 5). The beads were removed by centrifugation and the supernatant was recovered. The beads were further incubated with (100 mmol/L glycine pH 2.5, 1% Triton-X 100) to remove any protein not released by ATP hydrolysis (beads, see Fig 5). The proteins present in the various wash and elution supernatants were recovered by precipitation with 10% trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot. Immunodecorated proteins were detected with an antirabbit Ig-horseradish peroxidase conjugate and the Enhanced Chemiluminscent System (Pierce).
Immunofluorescence studies.
Sterile coverslips were preincubated with 5 μg/mL laminin overnight at 37°C. HEK-293 cells were seeded onto the laminin-coated coverslips and allowed to grow overnight. HEK-293 cells were washed once with PBS pH 7.0 containing 1.26 mmol/L CaCl2, 0.49 mmol/L MgCl2, 0.41 mmol/L MgSO4 (Ca/Mg PBS) and fixed with methanol for 6 minutes on dry ice. The cells were blocked with 5% fetal calf serum and 5% goat serum diluted in PBS (block buffer). Primary antibodies were diluted in block buffer and incubated with HEK-293 cells for 40 minutes. Cells were washed with the block buffer five times and incubated with either fluorescein isothiocyanate (FITC)-conjugated or Texas Red conjugated secondary antibodies (Vector Laboratories, Burlingame, CA) for 20 minutes. Cells were washed with block buffer five times, then twice with PBS. Cells were mounted on slides using Vectashield mounting medium (Vector Laboratories). Cells were examined under appropriate illumination with an E-600 epifluorescence microscope (Nikon, Melville, NY) with a 100X oil objective. Images were recorded with a U-III camera system (Nikon), and overlapping of the FITC and Texas Red images was done with Photoshop 5.0 (Adobe, San Jose, CA).
Resting platelets were prepared by adding 300 nmol/L prostaglandin I2 (Sigma). Platelets were separated from red blood cells by centrifugation at 150xg, collected by centrifugation at 900xg and resuspended in (140 mmol/L NaCl, 10 mmol/L NaHCO3, 2.5 mmol/L KCl, 0.5 mmol/L Na2HPO4, 1.0 mmol/L MgCl2, 22.0 mmol/L Na3 citrate, 0.55 mmol/L glucose, 0.35% bovine serum albumin [BSA], pH 6.5) with 25 μg/mL apyrase (Sigma) and 500 μmol/L CaCl2 and incubated for 15 minutes at 37°C. The platelets were seeded onto poly-lysine coated slides. Platelets were fixed in 3.7% formaldehyde (Sigma) for 20 minutes at RT and permeabilized with 100 μg/mL digitonin (Calbiochem, La Jolla, CA) for 5 minutes at RT. The remainder of the staining procedure was performed as above.
Platelets for surface activation were prepared without prostaglandin I2 or apyrase. Platelets were resuspended in PBS and seeded onto poly-lysine–coated slides. Platelets were either incubated with 10 μmol/L CaCl2 in PBS for 15 minutes or 10 μmol/L CaCl2 in PBS for 15 minutes followed by 0.1 U/mL thrombin (Sigma), 10 μmol/L CaCl2 in PBS for 15 minutes at RT. The remainder of the procedure was performed as above. Bone marrow samples containing megakaryocytes were seeded onto poly-lysine–coated slides, fixed in 3.7% formaldehyde, and permeabilized in 100 μmol/L digitonin. Cells were processed as above.
RESULTS
Cloning and characterizing human cellubrevin cDNA.
Degenerate PCR primers based on homologous regions of six previously cloned VAMPs were used to amplify new VAMP-encoding cDNAs from a megakaryocyte cDNA library. Primary screening of pools from a megakaryocyte-like CMK cell cDNA library yielded a 209-bp PCR product that was 88%, 79%, and 82% identical to the nucleotide sequences of rat cellubrevin, human VAMP 1, and 2, respectively. The one positive pool was then screened by colony hybridization using a riboprobe made from the initial PCR product. A single positive clone was isolated and sequenced (Fig 1A). The clone, termed human cellubrevin (Hceb), is 693 bp in length and contains: 24 bp of 5′ untranslated sequence, including an upstream stop codon; a 300-bp coding region; and 369 bp of 3′ untranslated sequence that does not include a poly A tract (Fig 1A). The nucleotide sequence immediately adjacent to the proposed initiator ATG is noncanonical (gccaaaATGt), yet it is identical to that found in rat cellubrevin.30
Nucleotide and predicted amino acid sequence of human cellubrevin. (A) Nucleotide and amino acid sequence of Hceb. The numbers correspond to the number of nucleotides and the underlined regions correspond to the sequences for which degenerate primers were designed. The sequence of the Hceb clone was submitted to Genbank, accession number U64520. (B) Comparison of the Hceb amino acid sequence to other human VAMPs and rat cellubrevin. The dots represent identical amino acids. The underlined region represents the peptide that was used to make a specific antihuman cellubrevin Ab. The bold methionine (M) represents the VAMP 2 vesicle targeting signal, which is also present in Hceb.
Nucleotide and predicted amino acid sequence of human cellubrevin. (A) Nucleotide and amino acid sequence of Hceb. The numbers correspond to the number of nucleotides and the underlined regions correspond to the sequences for which degenerate primers were designed. The sequence of the Hceb clone was submitted to Genbank, accession number U64520. (B) Comparison of the Hceb amino acid sequence to other human VAMPs and rat cellubrevin. The dots represent identical amino acids. The underlined region represents the peptide that was used to make a specific antihuman cellubrevin Ab. The bold methionine (M) represents the VAMP 2 vesicle targeting signal, which is also present in Hceb.
Human cellubrevin protein has an overall 92% amino acid identity to rat cellubrevin, 74% to human VAMP 1, and 83% to human VAMP 2 (alignment shown in Fig 1B). The homology is highest in the central core region (a.a.11-77), which is predicted to contain the two coiled-coil domains important for t-SNARE binding.15,16 In addition, Hceb contains the methionine 46 of VAMP 2, shown to be critical for synaptic vesicle targeting.14 This region also contains the specific cleavage sites used by tetanus and botulinum B, D, F, and G neurotoxins.33 Like other VAMPs, the N-terminal domain of Hceb is unique and there is only limited homology in the C-terminal transmembrane domain.
Human cellubrevin is widely distributed.
A Northern blot of poly A+ mRNA from the indicated human tissues was incubated with a [32P]-labeled nick translated probe made from 292 bp of the 3′ untranslated region of Hceb (bp 402-693 Fig 1A). mRNA load controls were confirmed with a [32P] nick translated β-actin DNA probe that will hybridize to more than one isoform in some tissues (pancreas and placenta) (Fig 2). The results of this Northern blot show that the Hceb message is expressed in every human tissue tested. The 2.5-kB message is similar to the size of VAMP 1 and 2 mRNAs34 and slightly larger than the message for rat cellubrevin (1.8 kB).30 Homologous expressed sequence tags (ESTs) derived from human lung, placenta, pancreas, pineal gland, pituitary gland, brain, and heart are available, but as listed in the database, none contain the entire coding region.
Human cellubrevin is expressed in many human tissues. The Northern blot containing 2 μg of poly A+ mRNA from the indicated human tissues was incubated with a [32P] nick-translated DNA probe made from the 292-bp of the 3′ untranslated region of human cellubrevin (bp 402-693, Fig 1A). The blot was washed and exposed to film for 18 hours at −80°C with an intensifying screen. The positions of the size standards in kilobases are shown at right. The same blot was also probed with a [32P] nick-translated β-actin DNA probe, washed, and exposed to film at −80°C for 2 hours. Some tissues contain two isoforms that will hybridize with the β-actin probe (pancreas and placenta).
Human cellubrevin is expressed in many human tissues. The Northern blot containing 2 μg of poly A+ mRNA from the indicated human tissues was incubated with a [32P] nick-translated DNA probe made from the 292-bp of the 3′ untranslated region of human cellubrevin (bp 402-693, Fig 1A). The blot was washed and exposed to film for 18 hours at −80°C with an intensifying screen. The positions of the size standards in kilobases are shown at right. The same blot was also probed with a [32P] nick-translated β-actin DNA probe, washed, and exposed to film at −80°C for 2 hours. Some tissues contain two isoforms that will hybridize with the β-actin probe (pancreas and placenta).
Antihuman cellubrevin antibody production and characterization.
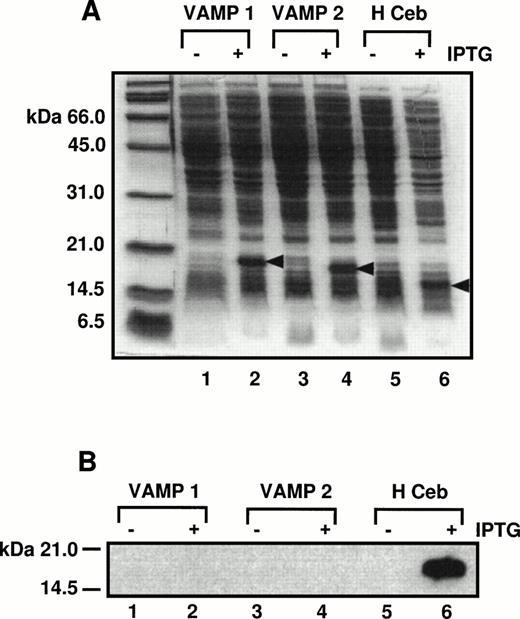
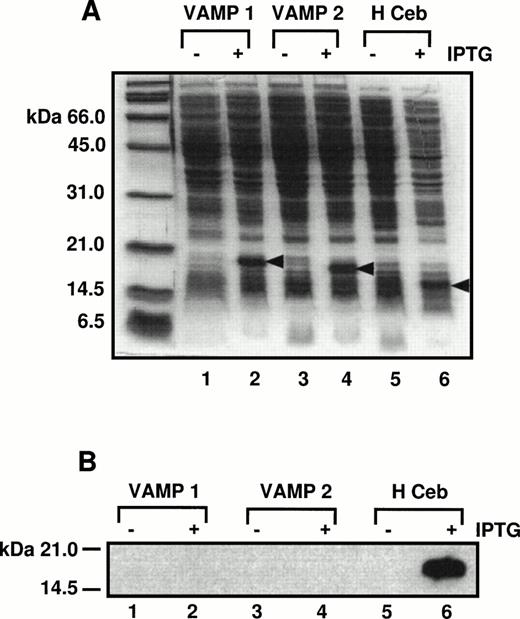
A specific antipeptide human cellubrevin antibody was made against the first 12 amino acids of the N-terminus of human cellubrevin. To show that this antibody is specific for Hceb, its reactivity against VAMP 1 and 2 was tested. E. coli cells expressing His6-tagged VAMP 1, 2, and Hceb constructs were induced with 1 mmol/L isopropyl thio-β-D-galactoside (IPTG) for 4 hours to produce each recombinant protein. The bacterial extracts were separated on a SDS-PAGE gel and stained with Coommassie Brilliant Blue dye (Fig 3A). The identical samples were separated on a SDS-PAGE gel, transferred to nitrocellulose, and probed with the anti-Hceb Ab (Fig 3B). As shown, the antipeptide antibody while detecting Hceb does not cross-react with either human VAMP 1 or 2.
Antihuman cellubrevin antibody does not cross-react with VAMP 1 or VAMP 2. (A) E. coli cells expressing His6-tagged VAMP 1, 2, and Hceb constructs were induced with 1 mmol/L IPTG for 4 hours to produce each recombinant protein. Bacterial extracts from the minus and plus IPTG-induced samples were separated on a 12.5% SDS-PAGE gel and proteins were stained with Coomassie Brilliant Blue. Lanes 1 and 2, VAMP 1 ± IPTG; lanes 3 and 4, VAMP 2 ± IPTG; lanes 5 and 6, Hceb ± IPTG. (B) The same samples were separated by SDS-PAGE and then transferred to nitrocellulose and blotted with a specific anti-Hceb Ab at a concentration of 1/1,000.
Antihuman cellubrevin antibody does not cross-react with VAMP 1 or VAMP 2. (A) E. coli cells expressing His6-tagged VAMP 1, 2, and Hceb constructs were induced with 1 mmol/L IPTG for 4 hours to produce each recombinant protein. Bacterial extracts from the minus and plus IPTG-induced samples were separated on a 12.5% SDS-PAGE gel and proteins were stained with Coomassie Brilliant Blue. Lanes 1 and 2, VAMP 1 ± IPTG; lanes 3 and 4, VAMP 2 ± IPTG; lanes 5 and 6, Hceb ± IPTG. (B) The same samples were separated by SDS-PAGE and then transferred to nitrocellulose and blotted with a specific anti-Hceb Ab at a concentration of 1/1,000.
Human cellubrevin colocalizes with the transferrin receptor in HEK-293 cells.
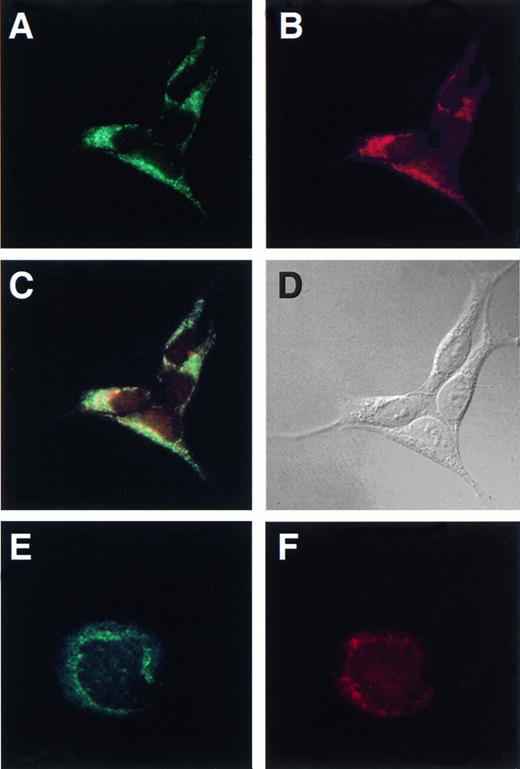
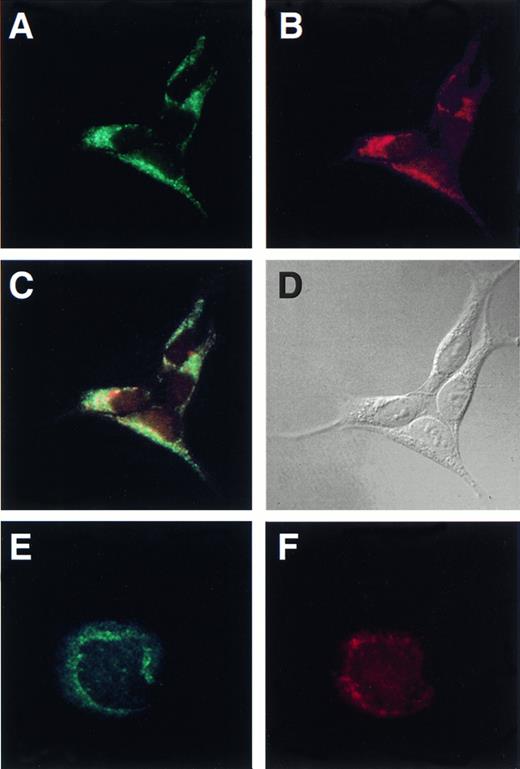
To determine the subcellular distribution of Hceb, we performed immunofluorescence microscopy with the anti-Hceb antibody on HEK-293 cells. HEK-293 cells showed a perinuclear punctate staining pattern with anti-Hceb Abs that did not resemble Golgi complex or endoplasmic reticulum staining (Fig 4A). This staining pattern appears similar to the antirat cellubrevin perinuclear, punctate staining of CHO cells.28 The transferrin receptor is often used as a marker for recycling endosomal compartments35 and was shown to colocalize with rodent cellubrevin in Chinese hamster ovary (CHO) cells. Double-labeling experiments in HEK-293 cells with anti-Hceb antibody and the antitransferrin receptor antibody were performed to determine if Hceb like rat cellubrevin, is localized to recycling endosomes. As shown in Fig 4A through C, the transferrin receptor and Hceb share a similar staining pattern, supporting the conclusion that Hceb is endosomal. When compared directly, the staining of transferrin receptor and Hceb showed a high degree of overlap as indicated in yellow (Fig4C). A similar pattern of anti-Hceb staining and overlap with transferrin receptor was observed in HepG2 cells (data not shown). The staining pattern of anti-lysosome-associated membrane protein (LAMP) 1 Ab, a late endosome and lysosomal marker, did not overlap with Hceb staining in HEK-293 and HepG2 cells (data not shown). These data are consistent with Hceb being the equivalent to rodent cellubrevin.
Human cellubrevin colocalizes with the transferrin receptor in HEK-293 cells and is present in megakaryocytes. HEK-293 cells were fixed with methanol and incubated with (A) anti-Hceb antibody (1/25), FITC-conjugated antirabbit antibody (1/150); (B) antitransferrin receptor antibody (1/25), Texas Red-conjugated antimouse antibody (1/150); (C) overlap of (A) and (B), which was generated with Adobe Photoshop 5.0; (D) differential interference contrast microscopy (DIC) image of the same cells. Original magnification 600×. Megakaryocytes were fixed with 3.7% formaldehyde, permeabilized with 100 μg/mL digitonin and incubated with (E) anti-Hceb antibody (1/25), FITC-conjugated antirabbit antibody (1/150); (F) anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150). Original magnification 1,000×.
Human cellubrevin colocalizes with the transferrin receptor in HEK-293 cells and is present in megakaryocytes. HEK-293 cells were fixed with methanol and incubated with (A) anti-Hceb antibody (1/25), FITC-conjugated antirabbit antibody (1/150); (B) antitransferrin receptor antibody (1/25), Texas Red-conjugated antimouse antibody (1/150); (C) overlap of (A) and (B), which was generated with Adobe Photoshop 5.0; (D) differential interference contrast microscopy (DIC) image of the same cells. Original magnification 600×. Megakaryocytes were fixed with 3.7% formaldehyde, permeabilized with 100 μg/mL digitonin and incubated with (E) anti-Hceb antibody (1/25), FITC-conjugated antirabbit antibody (1/150); (F) anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150). Original magnification 1,000×.
Human cellubrevin acts as a SNARE in platelets.
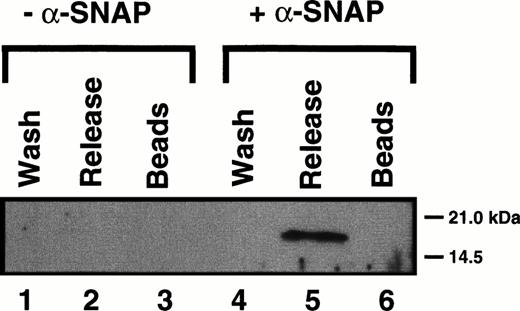
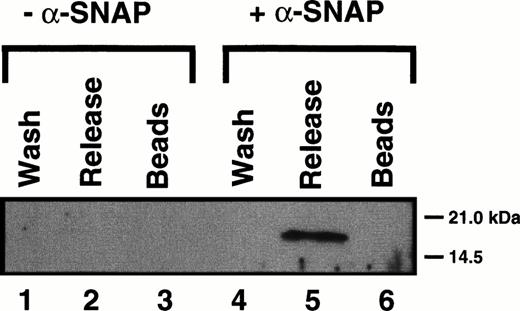
Having shown that Hceb exhibits properties consistent with it being the human homologue of rat cellubrevin, we next sought to determine if it was in fact present in platelets. Anti-Hceb antibody was able to detect a 16-kD protein in as little as 60 μg of salt-washed, detergent-solubilized platelet membrane extracts (data not shown). To show that Hceb in platelets has SNARE activity, we used the method of Söllner et al.17 HPE (5 mg), which contains all of the platelet SNARE molecules, was incubated with recombinant α-SNAP, γ-SNAP, and NSFmyc in the presence of EDTA and ATPγS. The recombinant proteins will interact with all of the platelet SNARE proteins to form 20S complexes, which can then be isolated by coimmunoprecipitation through the epitope-tagged NSF in the complex (Fig 5). The 20S complexes are washed and then incubated with ATP/Mg2+ to promote 20S complex disassembly. The proteins from the ATPγS/Mg2+ wash (wash, lanes 1 and 4), the ATP/Mg2+ release supernatant (release, lanes 2 and 5), and the glycine eluate (beads, lanes 3 and 6) were analyzed by Western blotting using the anti-Hceb antibody. As shown in Fig 5, Hceb participated in 20S complex formation, but only when the adapter protein α-SNAP was added (lanes 4 through 6). This SNAP-dependent complex formation together with the ATP/Mg2+-dependent release from NSF (lane 5) are characteristic of a SNARE protein as originally described.17,22 This data indicates that Hceb is an integral membrane protein present in platelets. It further shows that its t-SNARE partners must also be present because v-SNAREs alone will not associate with α-SNAP and NSF.36 As further proof of Hceb’s SNARE activity, when HPE was incubated with an α-SNAP–GST fusion protein, Hceb was precipitated with the α-SNAP–GST containing complexes (data not shown). To date, no proteins detectable by antibodies to rat cellubrevin, VAMP 1, or VAMP 2 have been observed using either purification technique31 (and data not shown).
Human cellubrevin participates in 20S particle assembly and ATP-dependent disassembly. Five milligrams of HPE were incubated with NSFmyc and either with (+) or without (-) recombinant-SNAP. 20S Complex was immunoprecipitated with an anti-myc antibody cross-linked to agarose beads. The beads were harvested by centrifugation, washed in the presence of ATPγS (wash), and then incubated with ATP/Mg. The resulting supernatant was harvested (release), and the beads were then stripped with glycine (beads). The proteins were recovered by trichloroacetic acid (TCA) precipitation and analyzed by Western blotting with an anti-Hceb antibody (1/100).
Human cellubrevin participates in 20S particle assembly and ATP-dependent disassembly. Five milligrams of HPE were incubated with NSFmyc and either with (+) or without (-) recombinant-SNAP. 20S Complex was immunoprecipitated with an anti-myc antibody cross-linked to agarose beads. The beads were harvested by centrifugation, washed in the presence of ATPγS (wash), and then incubated with ATP/Mg. The resulting supernatant was harvested (release), and the beads were then stripped with glycine (beads). The proteins were recovered by trichloroacetic acid (TCA) precipitation and analyzed by Western blotting with an anti-Hceb antibody (1/100).
Human cellubrevin exhibits punctate staining in platelets and megakaryocytes.
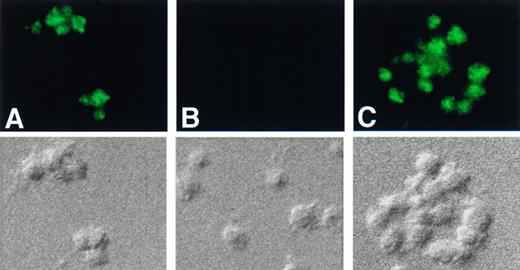
Immunofluorescence microscopy studies of platelets using the anti-Hceb antibody resulted in a punctate staining pattern in all platelets (Fig 6A). This pattern suggests that Hceb resides on an intracellular compartment, however, it did not overlap with the staining pattern seen using antibodies to other platelet granule markers (ie, LAMP-1, serotonin, von Willebrand factor [vWF]; data not shown). The anti-Hceb staining pattern was specific in that preincubation with recombinant Hceb eliminated immunoreactivity, whereas preincubation with recombinant VAMP 2 was without effect (Fig6B and C).
Immunofluorescence studies of human cellubrevin in platelets. Platelets were fixed with 3.7% formaldehyde and permeabilized with 100 μg/mL digitonin. The cells were then incubated with (A) anti-Hceb antibody (1/5) followed by FITC-conjugated antirabbit antibody (1/150); (B) anti-Hceb antibody that had been preincubated with 200 μg of GST-Hceb recombinant protein followed by FITC-conjugated antirabbit antibody (1/150); (C) anti-Hceb antibody that had been preincubated with 200 μg of GST-VAMP 2 recombinant protein followed by FITC-conjugated antirabbit antibody (1/150). A DIC image of each panel is included below. Original magnification 2,800×.
Immunofluorescence studies of human cellubrevin in platelets. Platelets were fixed with 3.7% formaldehyde and permeabilized with 100 μg/mL digitonin. The cells were then incubated with (A) anti-Hceb antibody (1/5) followed by FITC-conjugated antirabbit antibody (1/150); (B) anti-Hceb antibody that had been preincubated with 200 μg of GST-Hceb recombinant protein followed by FITC-conjugated antirabbit antibody (1/150); (C) anti-Hceb antibody that had been preincubated with 200 μg of GST-VAMP 2 recombinant protein followed by FITC-conjugated antirabbit antibody (1/150). A DIC image of each panel is included below. Original magnification 2,800×.
To determine if Hceb was also expressed in the megakaryocyte, we performed double-labeling, immunofluorescence microscopy on bone marrow megakaryocytes with a megakaryocytic marker antibody against gpIIβ/IIIα,37 (Fig 4E) and the anti-Hceb Ab (Fig 4F). The double positive staining allows identification of the mature megakaryocyte and shows that Hceb is present not only in platelets, but also in their bone marrow progenitor cell. Hceb shows punctate perinuclear staining that is similar to, but does not completely overlap with the fibrinogen receptor staining.
Hceb staining remains intracellular in surface-activated platelets.
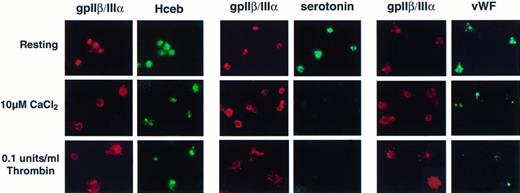
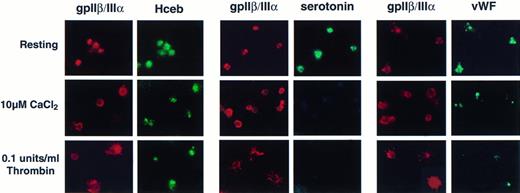
To determine if Hceb was mobilized to the plasma membrane on stimulation, platelets were seeded onto poly-lysine–coated slides and either maintained in a resting state, surface-stimulated in the presence of 10 μmol/L CaCl2, or stimulated with 0.1 U/mL thrombin. The distribution of the fibrinogen receptor (gpIIβ/IIIα) can be used as a marker to distinguish resting platelets from the various states of platelet activation.38-40 In resting platelets, the antifibrinogen receptor Ab shows an internal punctate staining pattern (Fig 7, first row). When surface-stimulated in the presence of CaCl2, the platelets displayed a distinct ring-like structure (Fig 7, middle row). In thrombin stimulated platelets, spikes or finger-like pseudopodia were clearly delineated by the antifibrinogen receptor staining (Fig 7, bottom row) indicating mobilization of gpIIβ/IIIα to the cell surface. The final, completely spread stage of surface-activated platelets39 is not represented. In the left panel, the platelets are doubled-stained with an anti-gpIIβ/IIIα Ab and an anti-Hceb Ab. The Hceb staining is punctate in resting platelets and remains internal, but centrally concentrated in stimulated platelets. In the middle panel of Fig 7, platelets were double-stained for serotonin and the fibrinogen receptor. The antiserotonin Ab staining is internally punctate in resting platelets and is completely abolished in stimulated platelets, suggesting that the dense core granules contents have been released. In the right most panel, platelets were double-stained for fibrinogen receptor and vWF. Again, resting platelets exhibit a granular staining pattern, but the stimulated platelets lost most, if not all, of the vWF staining. The Hceb positive organelle remains detectable in activated platelets even after serotonin and vWF immunoreactivity have all but disappeared. Interestingly, Hceb, unlike the fibrinogen receptor, does not mobilize to the cell surface after stimulation.
Human cellubrevin staining remains intracellular in surface-activated platelets. Platelets were prepared as resting (see Materials and Methods) or surface-stimulated in the presence of 10 μmol/L CaCl2 or 10 μmol/L CaCl2 and 0.1 U/mL thrombin. Platelets were fixed with 3.7% formaldehyde and permeabilized with 100 μg/mL digitonin. Left panel, anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150) (gpIIβ/III); anti-Hceb antibody (1/25), FITC-conjugated antirabbit antibody (1/150) (Hceb). Middle panel, anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150) (gpIIβ/III); antiserotonin antibody (1/10), FITC-conjugated antirabbit antibody (1/150) (serotonin). Right panel, anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150) (gpIIβ/III); anti-vWF antibody (1-500), FITC-conjugated antirabbit antibody (1/500) (vWF). Original magnification 2,800×.
Human cellubrevin staining remains intracellular in surface-activated platelets. Platelets were prepared as resting (see Materials and Methods) or surface-stimulated in the presence of 10 μmol/L CaCl2 or 10 μmol/L CaCl2 and 0.1 U/mL thrombin. Platelets were fixed with 3.7% formaldehyde and permeabilized with 100 μg/mL digitonin. Left panel, anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150) (gpIIβ/III); anti-Hceb antibody (1/25), FITC-conjugated antirabbit antibody (1/150) (Hceb). Middle panel, anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150) (gpIIβ/III); antiserotonin antibody (1/10), FITC-conjugated antirabbit antibody (1/150) (serotonin). Right panel, anti-gpIIβ/III antibody (1/100), Texas Red-conjugated antimouse antibody (1/150) (gpIIβ/III); anti-vWF antibody (1-500), FITC-conjugated antirabbit antibody (1/500) (vWF). Original magnification 2,800×.
DISCUSSION
In an attempt to identify VAMP-like molecules in platelets, we screened a megakaryocyte cDNA library with degenerate PCR primers based on conserved VAMP sequences. We isolated one clone, termed human cellubrevin, which is highly homologous to other VAMP proteins except for its divergent N-terminus. Hceb mRNA is widely expressed, and the protein is present in each human cell type tested; HEK-293 cells (kidney), HepG2 (liver), bone marrow megakaryocytes, and platelets. In HEK-293 cells, Hceb has similar staining pattern to the transferrin receptor, supporting the proposal that human cellubrevin is functioning in an endocytic pathway. In platelets, Hceb is an integral membrane protein that participates in 20S complex formation and ATP-dependent disassembly demonstrating its function as a SNARE protein. This also indicates that its t-SNARE partners are present, as v-SNAREs need t-SNAREs to participate in 20S complex formation.36
Immunofluorescence studies with an anti-Hceb antibody showed a punctate, intracellular staining pattern in resting, as well as in stimulated platelets (Fig 7). In contrast, antibodies to serotonin (dense core granule) and vWF (alpha granule) showed a punctate staining pattern in resting cells that all but disappeared when the cells were stimulated. Compared with the membrane protein gpIIβ/IIIα, Hceb does not mobilize to the cell surface on stimulation. Two possible scenarios could explain these results. Hceb could be present on a granule that is involved in exocytosis, but is somehow trapped in the granule membrane and prevented from laterally diffusing to the plasma membrane after fusion. Alternatively, Hceb could be present on an organelle that is not involved in exocytosis. This later possibility is in keeping with preliminary immunofluorescence studies that show only limited overlap between Hceb positive compartments and those that stain with antibodies to serotonin, vWF, or LAMP-1 (Bernstein and Whiteheart unpublished). Further, immunoelectron microscopy studies will be required to more definitively define Hceb’s intraplatelet locale.
Studies of the rodent cellubrevin in nucleated cells suggest that it plays a role in endocytic pathways. Tetanus toxin cleaves and inactivates cellubrevin.30 When added to streptolysin O-permeabilized CHO cells, tetanus toxin causes a 30% decrease in the recycling of preloaded [125I]-labeled transferrin.28 Studies of the role that cellubrevin plays in Glut 4 mobilization have been more equivocal and seem to depend on the mode of introducing the toxin. In the best case, when adipocytes were incubated overnight with toxin in isotonic, low ionic strength media, there was a 64% decrease in the insulin-stimulated glucose uptake.41 At this stage, it appears that cellubrevin does play a role in the overall process of receptor recycling, yet it is unclear at which transport step it functions.
Platelets and megakaryocytes undergo endocytosis.42Clathrin-coated vesicles containing fibrinogen bud from the OCS and fuse with alpha granules, loading them with fibrinogen.43Recently, endocytic compartments termed MVB I and II have been defined in platelets and megakaryocytes.8 Kinetic uptake experiments performed in megakaryocytes showed a sequential uptake of BSA and fibrinogen into the MVBs and then into alpha granules. After granule-granule fusion, the fibrinogen receptor (gpIIβ/IIIα) may be recycled back to the plasma membrane.44 Consistent with its proposed role in other cells, Hceb could function in this or other endocytic pathways in megakaryocytes and platelets. If Hceb is involved in the endocytic process, it is likely most active in megakaryocytes when loading of alpha granules is most prevalent. However, based on the apparent abundance of Hceb in platelets, its homogeneous expression in all platelets examined and the fact that its t-SNARE partners are also present in platelet membrane extracts, Hceb could still function in platelets. Further studies will be required to determine the role of this v-SNARE in platelets and megakaryocytes.
ACKNOWLEDGMENT
We would like to thank Dr Takeyuki Sato for providing the CMK cDNA plasmid library, Dr Reinhard Jahn for the anti-VAMP 1 and anti-VAMP 2 Ab (C1 69.1) antibodies, Dr P. DeCamilli for the antirat cellubrevin antibody, Dr G. Vanzant for the bone marrow samples, and the staff of the Kentucky Blood Center for their assistance. We would also like to thank the members of the Whiteheart laboratory for their productive discussions and help.
Supported by Grant No. CB-153 from the American Cancer Society (to S.W.W.) and Grant No. HL56652 from the National Institutes of Health (to S.W.W.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
NOTE ADDED IN PROOF
During the revision of this manuscript, a publication by Galli et al (Mol Biol Cell 9:1437,1998) appeared. In this report, the investigators concurred with the conclusion that our clone represents the human homologue of the rodent cellubrevin. The Genbank submission (U64520) was referenced in their text.
Author notes
Address reprint requests to Sidney W. Whiteheart, PhD, Department of Biochemistry, University of Kentucky College of Medicine, Chandler Medical Center, 800 Rose St, Lexington, KY 40536-0084; e-mail:whitehe@pop.uky.edu.



![Fig. 2. Human cellubrevin is expressed in many human tissues. The Northern blot containing 2 μg of poly A+ mRNA from the indicated human tissues was incubated with a [32P] nick-translated DNA probe made from the 292-bp of the 3′ untranslated region of human cellubrevin (bp 402-693, Fig 1A). The blot was washed and exposed to film for 18 hours at −80°C with an intensifying screen. The positions of the size standards in kilobases are shown at right. The same blot was also probed with a [32P] nick-translated β-actin DNA probe, washed, and exposed to film at −80°C for 2 hours. Some tissues contain two isoforms that will hybridize with the β-actin probe (pancreas and placenta).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/2/10.1182_blood.v93.2.571/4/m_blod40205002w.jpeg?Expires=1763676844&Signature=B7~BJzaH0aUOvsAhsC~XQCFZe2cdpec2-b4PDGK3B-2CFJLoQ7ztNhNOacBlh-kGsVm2-lpehUVQCg6MgNDs9xWpuLJDMhY-7XgGtHavPXDrj0C42JFBpjsumax60YTWYyWfg~pytsaY9TlckHtjayjJOPQECiI6nDEK5yv9JkkeyzWQJHgtoKwJ1YhbRlR6tox8A0UdyScyYGHh2NUe~JU1fHsIcE10v5KR5FZgCD5dcPCsHCQAFtqQPxgCXH9H5IHUUUyfmp6JJrRz~pMqd4P3WdEi2cFoqlbcrJP4fYs6yOgswihPfeLRA3VNNRwFPAUUx~w30CWXVRs0YkZAgQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Fig. 2. Human cellubrevin is expressed in many human tissues. The Northern blot containing 2 μg of poly A+ mRNA from the indicated human tissues was incubated with a [32P] nick-translated DNA probe made from the 292-bp of the 3′ untranslated region of human cellubrevin (bp 402-693, Fig 1A). The blot was washed and exposed to film for 18 hours at −80°C with an intensifying screen. The positions of the size standards in kilobases are shown at right. The same blot was also probed with a [32P] nick-translated β-actin DNA probe, washed, and exposed to film at −80°C for 2 hours. Some tissues contain two isoforms that will hybridize with the β-actin probe (pancreas and placenta).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/2/10.1182_blood.v93.2.571/4/m_blod40205002w.jpeg?Expires=1763676845&Signature=MOvx4io0hYsfFOJYsqCqXg-wVHXA3oFUS~8Scx5KRHE~~7KANGfOZfMsfqLKrrpofnm283BAH~qi3SXTbH-Q9qhhDq53Jqm0tdrC5uG83Vn1YXB2KAYUJ-V9CIULM4AfNyZjY6iMuhzWmHeVWi5EpGxsdyQyb19EQIQW3-dwn1vupGwPthiWLGse1bTq2i5Q5kHDG-g40Vm7aZ8rZZe2VP4pL~4kWk1vrOfKL1jypXpbSiIzdEbZ2P3~fXYRRRZdHPEkFkQJeXoSM3sXT4-w8ZbOb~wqjZEOqmZw1lyt1i5FpXfSYCl7-V0jKFvrl-6OQDTOKq-D1JqAsjPcZS8QZg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)