To the Editor:
Fanconi anemia (FA) is an autosomal recessive disease characterized by bone marrow failure and cancer susceptibility. At least eight complementation groups for FA have been identified, and the genes corresponding to groups A, C, and G have been cloned. An understanding of the biochemical function of the FANCA, FANCC, and FANCG proteins would yield considerable insight to the cellular function of the FA pathway. Unfortunately, the three cloned FA proteins have no recognizable structural or functional motifs, and studies regarding their interaction and cellular localization in most cases have been conflicting. First, some studies show that the FANCA and FANCC protein bind in a functional complex,1,2 while other studies fail to detect the complex.3 Second, some studies show that the FANCC protein is localized to the cytoplasm and nucleus,1,4 while other studies show that FANCC is primarily cytoplasmic.5
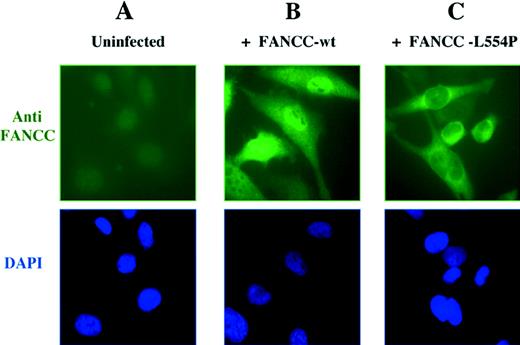
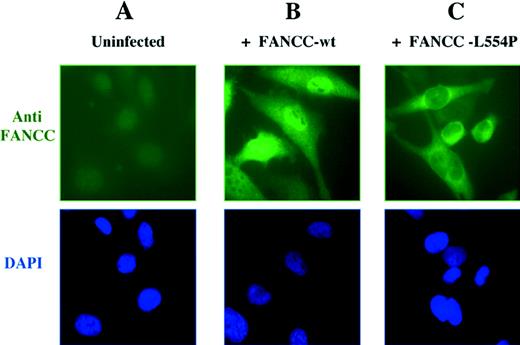
To resolve this controversy, Hoatlin et al6 have used confocal microscopy to determine that FANCC is localized to both cytoplasmic and nuclear compartments. We have new evidence that strongly supports this model of Hoatlin et al. We analyzed the subcellular localization of FANCC polypeptide in retrovirally infected HeLa cells by immunofluorescence microscopy (Fig1). The wild-type FANCC protein was detected in both nucleus and cytoplasm in varying ratios (Fig 1B), confirming the results of Hoatlin et al.6 In most cells, the protein was predominantly localized to the nucleus. In a small percentage of cells, a strong cytoplasmic localization of FANCC was also detected. An examination of the cell morphology and DAPI staining showed that these cells expressing cytoplasmic FANCC were undergoing mitosis or had just completed cell division, suggesting that localization may be cell-cycle dependent.
Cellular localization of the wild-type FANCC protein and the L554P mutant. (A) The HeLa cell line expresses low levels of endogenous FANCC protein that is undetectable by anti-FANCC immunofluorescence. HeLa cells were infected with retroviral supernatants, pMMP-FANCC (B) or pMMP-FANCC-L554P (C). Pools of infected cells were stained with anti FANCC and the DNA-specific dye, DAPI (4′,6-diamidino-2-phenylindole) (bottom images) and analyzed by immunofluorescence, as previously described.4
Cellular localization of the wild-type FANCC protein and the L554P mutant. (A) The HeLa cell line expresses low levels of endogenous FANCC protein that is undetectable by anti-FANCC immunofluorescence. HeLa cells were infected with retroviral supernatants, pMMP-FANCC (B) or pMMP-FANCC-L554P (C). Pools of infected cells were stained with anti FANCC and the DNA-specific dye, DAPI (4′,6-diamidino-2-phenylindole) (bottom images) and analyzed by immunofluorescence, as previously described.4
Mutations in FA patients represent an important resource for understanding both the function of the FA proteins and the molecular basis of the disease. For instance, the FA patient-derived FANCC-L554P mutant, which has a leucine to proline mutation at amino acid 554, fails to complement the mitomycin-C sensitivity of FA-C cell lines7 and is defective in FANCA binding.1 To evaluate the effect of the L554P mutation on the subcellular localization of the protein, we analyzed retrovirally infected HeLa cells by indirect immunofluorescence. Interestingly, the FANCC-L554P mutant, unlike the wild-type protein, was only observed in the cytoplasm of retrovirally infected HeLa cells (Fig 1C).
On the basis of these results, we conclude that the wild-type FANCC protein is normally localized in both nucleus and cytoplasm. The absence of nuclear accumulation of the FANCC-L554P protein allows several conclusions. First, the nuclear localization of wild-type FANCC is not merely caused by the protein overexpression. Second, the functional activity of the FANCC protein requires nuclear localization. The FANCC-L554P mutation results in cytoplasmic localization and abolishes function. The missense mutation is likely to disrupt a domain of the protein required for its functional activity. The COOH-terminus domain of FANCC may be required for binding to FANCA or other protein subunits of the FA complex or for localization or stabilization of the protein in the nucleus.