Targeted expression to specific tissues or cell lineages is a necessary feature of a gene therapy vector for many clinical applications, such as correction of hemoglobinopathies or thalassemias by transplantation of genetically modified hematopoietic stem cells. We developed retroviral vectors in which the constitutive viral enhancer in the U3 region of the 3′ LTR is replaced by an autoregulatory enhancer of the erythroid-specific GATA-1 transcription factor gene. The replaced enhancer is propagated to the 5′ LTR upon integration into the target cell genome. The modified vectors were used to transduce human hematopoietic cell lines, cord blood-derived CD34+ stem/progenitor cells, and murine bone marrow repopulating stem cells. The expression of appropriate reporter genes (▵LNGFR, EGFP) was analyzed in the differentiated progeny of transduced stem cells in vitro, in liquid culture as well as in clonogenic assay, and in vivo, after bone marrow transplantation in lethally irradiated mice. The GATA-1 autoregulatory enhancer effectively restricts the expression of the LTR-driven proviral transcription unit to the erythroblastic progeny of both human progenitors and mouse-repopulating stem cells. Packaging of viral particles, integration into the target genome, and stability of the integrated provirus are not affected by the LTR modification. Enhancer replacement is therefore an effective strategy to target expression of a retroviral transgene to a specific progeny of transduced hematopoietic stem cells.
RETROVIRAL VECTORS are widely used to integrate and express exogenous genes into a variety of animal cells and provide a safe and relatively efficient gene transfer tool for human gene therapy applications.1-3 In some of these applications, controlling the expression of a transferred, therapeutic gene at quantitative and qualitative (tissue- or cell-specific) level is a critical, and often limiting, factor. The most frequently used backbone for the construction of retroviral vectors is the Moloney murine leukemia virus (MoMLV). Transcription from the MoMLV long terminal repeat (LTR) is dependent on the viral promoter/enhancer elements located in the U3 region of the 5′ LTR, which allows constitutive expression of a transferred gene in most cell types. Conversely, tissue-specific or inducible expression is difficult to obtain. Although an independently controlled transcriptional unit can be transferred and expressed in the framework of a retroviral vector, the strong activity of the MoMLV enhancer/promoter usually interferes with, or prevents, regulation and function of most eukaryoticcis-acting elements, even in the proper cell context.4-6 Alternative designs have been developed in an attempt to overcome this problem, from self-inactivating vectors, in which deletion of viral elements allows expression from an internal promoter,7-9 to vectors in which a transgene is transcribed in opposite orientation with respect to the viral transcription unit,10 to double-copy vectors in which a complete minigene is inserted into the LTR upstream from the U3 region.11These strategies frequently raise other problems, such as generation of antisense transcripts, proviral instability, or decreased viral titer. Attempts to redirect the promiscuous MoMLV LTR transcriptional activity by adding to or replacing the viral enhancer with heterologous control elements from other viruses or cellular genes have been partially successful, allowing investigators to change the vector tropism12 or increase transgene expression in specific tissues13 or cell contexts.14
For correction of genetic disorders such as hemoglobinopathies or thalassemias, in which the ultimate goal is to restrict transgene expression to a specific progeny—the erythropoietic compartment—of the hematopoietic stem cell, transcriptional targeting of the vector is a mandatory requirement. We report the development of targeted retroviral vectors obtained by replacing the LTR extended viral enhancer with an autoregulatory enhancer (ARE) of the erythroid-specific, zinc finger transcription factor GATA-1.15,16 The GATA-1 ARE is a 200-bp, conserved element upstream of the GATA-1 erythroid-specific promoter (−856 to −655) and contains two palyndromic GATA-1 binding sites. This element was previously shown to restrict transcription of a heterologous promoter to human or murine erythroblastic cell lines in transient transfection.17,18 The LTR modification was introduced in retroviral vectors encoding either a truncated form of the human p75, low-affinity nerve growth factor receptor (ΔLNGFR), or a humanized form of the green fluorescent protein (EGFP) to allow detection of transgene expression with single-cell resolution by flow cytometry19 or in situ in a colony-forming assay. Human cord blood-derived hematopoietic stem/progenitor cells, including the CD34+/CD38low fraction, and mouse repopulating bone marrow (BM) stem cells were transduced by the modified vectors and analyzed for transgene expression in the differentiated progenies in vitro or after BM transplantation ex vivo. We show that enhancer replacement does not affect transduction and stability of the retroviral vectors and effectively restricts LTR-driven expression to the erythropoietic progeny of transduced stem cells.
MATERIALS AND METHODS
Retroviral vectors and producer cell lines.
The SFCM retroviral vector, encoding the ΔLNGFR cDNA under the control of the viral LTR, was previously described.20 The ΔSFCM vector was derived by replacing the enhancer and CAAT box in the U3 region of the 3′ LTR, from the Nhe I site to position −60 from the transcription start site, with a 23-bp polylinker. The GATA-SFCM vector was obtained by cloning the 0.2-kbBamHI fragment of the pSV0GATA plasmid18 containing the GATA-1 ARE in the SnaBI site of the ΔSFCM LTR polylinker. The LGSN vector was constructed by cloning theEco47III/Sca I fragment of the pEGFP-C3 plasmid (Clontech, Palo Alto, CA) containing the EGFP gene in the Hpa I site of the LXSN vector.21 The GATA-LGSN vector was obtained by replacing the ΔLNGFR insert of GATA-SFCM with the EGFP insert of LGSN.
Packaging lines for LGSN and SFCM were generated by transinfection in the amphotropic GP+envAm1222 and ecotropic GP+E-8623 cells, as previously described,20whereas those for ΔSFCM, GATA-SFCM, and GATA-LGSN were obtained by plasmid transfection and selection in 0.8 mg/mL G418 (Boehringer Mannheim, Mannheim, Germany). Viral titers were assayed by transfer of G418 resistance to NIH-3T3 fibroblasts.
Transduction of hematopoietic cells.
Mouse erythroleukemia (MEL) cells were transduced as described.24 Human hematopoietic K562, Kasumi-1, U937, and MOLT-3 cell lines (ATCC, Rockville, MD) were grown in RPMI 1640 (GIBCO, Grand Island, NY) supplemented with 10% fetal calf serum (FCS), transduced by 48 hours of cocultivation with irradiated (100 Gy) packaging cells in the presence of polybrene (8 μg/mL), and selected as bulk cultures in 1.0 to 1.5 mg/mL of G418.
Human CD34+ cells were purified from the Ficoll (Lymphoprep; Nycomed Pharma, Oslo, Norway) mononuclear cell fraction of umbilical cord blood by a two-round separation procedure using the CD34 magnetic cell isolation kit (MiniMacs; Miltenyi, Auburn, CA). CD34+ cells (5 to 10 × 104) were cultured in Iscove’s modified Dulbecco’s medium (IMDM; GIBCO) containing 20% FCS, 10 ng/mL human interleukin-3 (huIL-3), huIL-6, and human stem cell factor (huSCF; all from R&D Systems, Minneapolis, MN) for 48 hours and transduced by multiple cycles of infection with retroviral supernatant in the presence of polybrene (4 μg/mL). Transduction was performed in the upper chamber of Transwell (Costar, Cambridge, MA) coculture plates above the murine, clonal stromal cell line CBR-BM, D#1.25 Cells were then cultured for an additional 48 hours in medium supplemented with 4 U/mL human erythropoietin (huEpo; Janssen-Cilag, Geel, Belgium).
Transduced CD34+ cells were grown in liquid culture in X-VIVO 20 serum-free medium (Biowhittaker, Walkersville, MD) supplemented with 10 ng/mL huSCF and 4 U/mL huEpo to induce erythroid differentiation or were grown in IMDM supplemented with 10% FCS, 10 ng/mL huSCF, huIL-6, huIL-3, and human granulocyte-macrophage colony-stimulating factor (huGM-CSF) to induce myeloid differentiation and were then analyzed by flow cytometry. Colony-forming assay was performed by plating 5 to 10 × 102 transduced CD34+ cells in 1 mL of methylcellulose culture medium (Stem Cell Technologies, Vancouver, British Columbia, Canada) containing 50 ng/mL huSCF, 10 ng/mL huGM-CSF, 10 ng/mL huIL-3, 3 U/mL huEpo, and 1.2 to 1.8 mg/mL G418. Burst-forming unit-erythroid (BFU-E) and colony-forming unit–granulocyte-macrophage (CFU-GM) colonies were scored 10 to 14 days after plating, harvested, and analyzed for ΔLNGFR expression by immunocytochemistry.
Analysis of transduced cells.
Expression of ΔLNGFR was monitored by flow cytometry (FACScan; Becton Dickinson, Mountain View, CA) using the murine antihuman p75-NGFR monoclonal antibody (MoAb) 20.4 (ATCC), conjugated to fluorescein isothiocyanate (FITC). Human cell surface phenotype was determined by flow cytometry using phycoerythrin (PE)-conjugated antihuman CD34 (Becton Dickinson), Glycophorin A (GpA) and CD13 (DAKO A/S, Glostrup, Denmark), and tricolor (TC)-conjugated antihuman CD38 (Becton Dickinson) MoAbs. Mouse cell phenotyping was performed using PE-conjugated antimouse Sca-1, TER-119, and Gr-1 (PharMingen, San Diego, CA) antibodies. The FITC-conjugated, antimouse CD45.1 MoAb (PharMingen) was used to evaluate donor-host chimerism in BM-transplanted mice. Isotype-matched nonspecific antibodies were used as controls.
Cyto-centrifuged cell preparations were acetone-fixed and analyzed for ΔLNGFR expression by immunocytochemistry with alkaline phosphatase antialkaline phosphatase (APAAP) complex following standard methods. Samples were incubated for 1 hour with 20.4 MoAb (1:50 dilution), washed, incubated for 30 minutes with rabbit antimouse IgG (DAKO A/S; 1:25 dilution), and incubated with APAAP complex (DAKO A/S; 1:50 dilution) for an additional 30 minutes. The reaction was developed in Tris-buffered saline, pH 8.2, containing Naphtol AS-MX, Fast Red TR salt, and levamisole (all from Sigma, St Louis, MO). Slides were counterstained with hematoxylin before analysis.
RNA analysis.
Total cellular RNA was extracted by guanidine-isothiocyanate, poly(A)+ selected by oligo(dT)-cellulose chromatography, size-fractionated on 1% agarose-formaldeyde gel, Northern blotted to nylon membranes, and hybridized to 107 dpm of [32P]-labeled probes for the neomycin phosphotransferase (NeoR), GATA-1,26 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes, as previously described.19
Transduction of murine BM and BM transplantation.
Murine BM cells were harvested from C57Bl/6-Ly-5.1 mice (B/6.SJL-CD45a-Pep3b; Jackson Laboratories, Bar Harbor, ME) by flushing femurs and tibiae; prestimulated for 24 hours in IMDM supplemented with 20% FCS, 10 ng/mL muSCF, muIL-3, and huIL-6 (all from R&D Systems); and infected by coculture for 72 hours with irradiated (100 Gy) ecotropic packaging cells in the presence of polybrene (4 μg/mL). Transduced cells were analyzed for efficiency of gene transfer by flow cytometry and injected (1.0 to 1.5 × 107 cells/mouse) into the tail vein of recipient 8-to 12-week-old male C57BL/6 mice (Charles River, Calco, Italy) irradiated with two split doses of 400 cGy 4 hours apart. Eight weeks after BM transplantation, animals were killed and hematopoietic tissues were collected for FACS and immunocytochemical analysis.
RESULTS
Targeting retroviral vectors by LTR enhancer replacement.
The GATA-1 ARE was inserted in the U3 region of the 3′ LTR of the previously described SFCM vector,20 in which the cDNA for the marker gene ΔLNGFR and a NeoR gene are under the control of the viral LTR and of an internal SV40 promoter, respectively (Fig 1B). The modification is transferred to the 5′ LTR upon reverse transcription and integration of the provirus, allowing the chimeric LTR to direct transcription of the ΔLNGFR transgene. We first replaced the LTR enhancer direct repeats (from position −419 to −149 from the transcription start site) with the GATA-1 ARE. The residual activity of this partially deleted LTR was still constitutively high in all tested cells, although reduced approximately fivefold as compared with a wild-type LTR (data not shown). We therefore generated the ΔSFCM vector by deleting the viral enhancer and CAAT box (from position −419 to −60), leaving intact only the TATA and initiator elements (Fig 1B). The GATA-SFCM vector was constructed by cloning the GATA-1 ARE in the LTR of ΔSFCM (Fig 1B). Retroviral vectors containing the EGFP gene as an alternative reporter (LGSN and GATA-LGSN) were built following the same strategy. Amphotropic and ecotropic producer cell lines were established by transfecting hybrid-LTR plasmid vectors and isolating G418-resistant clones.
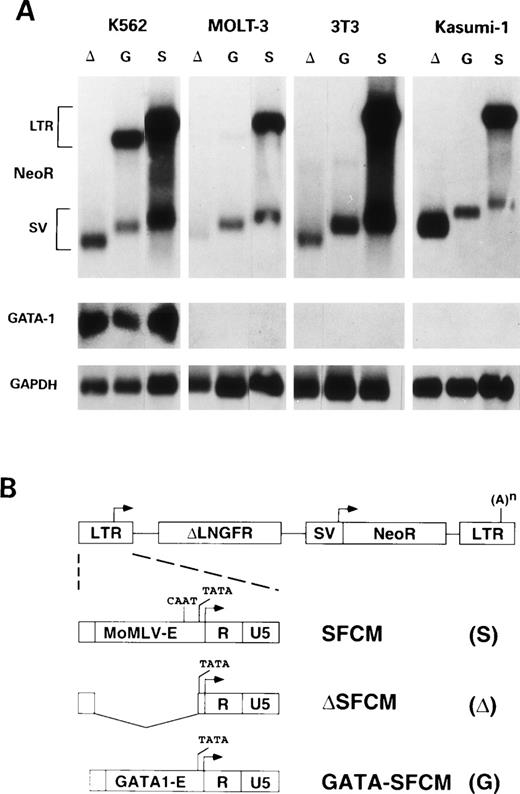
(A) Northern blot analysis of transcripts from K562 (erythroblastic), 3T3 (fibroblastic), MOLT-3 (T-lymphocytic), and Kasumi-1 (myeloblastic) cell lines transduced with ▵SFCM (lanes ▵), GATA-SFCM (lanes G), and SFCM (lanes S) retroviral vectors, after hybridization to NeoR (upper panels), GATA-1 (middle panels), or GAPDH (lower panels) probes. Transcripts from the viral LTR (LTR) or the internal SV40 (SV) promoters are indicated on the left. (B) Schematic representation of the SFCM retroviral provirus and 5′ LTR and of the modified 5′ LTRs of the ▵SFCM and GATA-SFCM vectors. LTR and SV40 promoters are indicated by arrows. (A)n indicates the polyadenylation site. The LTR U3 wild-type enhancer (MoMLV-E), the GATA-1 enhancer (GATA1-E), the LTR R and U5 regions, and the CAAT and TATA elements are indicated.
(A) Northern blot analysis of transcripts from K562 (erythroblastic), 3T3 (fibroblastic), MOLT-3 (T-lymphocytic), and Kasumi-1 (myeloblastic) cell lines transduced with ▵SFCM (lanes ▵), GATA-SFCM (lanes G), and SFCM (lanes S) retroviral vectors, after hybridization to NeoR (upper panels), GATA-1 (middle panels), or GAPDH (lower panels) probes. Transcripts from the viral LTR (LTR) or the internal SV40 (SV) promoters are indicated on the left. (B) Schematic representation of the SFCM retroviral provirus and 5′ LTR and of the modified 5′ LTRs of the ▵SFCM and GATA-SFCM vectors. LTR and SV40 promoters are indicated by arrows. (A)n indicates the polyadenylation site. The LTR U3 wild-type enhancer (MoMLV-E), the GATA-1 enhancer (GATA1-E), the LTR R and U5 regions, and the CAAT and TATA elements are indicated.
Efficiency of gene transfer was assayed by infecting GATA-1–expressing cell lines (human K562 for amphotropic and MEL cells for ecotropic vectors) and counting ΔLNGFR+ or EGFP+ cells by FACS analysis 48 hours after infection. Positive cells ranged from 5% to 25% in cultures transduced by enhancer-replaced LTR vectors and from 50% to 80% in cultures transduced by wild-type LTR vectors (data not shown). The differences correlated with the viral titers assayed on NIH/3T3 cells, which ranged from 104 to 105cfu/mL for ΔSFCM, GATA-SFCM, and GATA-LGSN vectors to 106cfu/mL for SFCM and LGSN vectors.
The GATA-SFCM vector is expressed only in erythroblastic cell lines.
The transcriptional activity of the chimeric LTR was tested in different cell lines of hemato-lymphopoietic origin. Human myeloblastic (Kasumi-1), monoblastic (U937), erythroblastic (K562), and T-lymphoblastic (MOLT-3) cell lines and murine erythroblastic (MEL) and fibroblasts (NIH 3T3) cells were transduced with the SFCM, ΔSFCM, and GATA-SFCM retroviral vectors and selected as bulk cultures for resistance to G418. Southern blot analysis of vector integration showed that modifications in the 3′ LTRs were correctly transferred to the 5′ LTRs, originating intact proviruses integrated at multiple sites in all target cell populations (data not shown). Expression of vector-specific transcripts was analyzed by Northern blotting of poly(A)+ RNA and hybridization to a NeoR-specific probe. LTR-driven transcripts were never detected in cells transduced with the ΔSFCM vector (Fig 1A, lanes Δ), indicating that deletion of viral enhancer/CAAT sequence virtually abrogated transcription from the LTR promoter. Genomic transcripts derived from the modified LTR of the GATA-SFCM vector were detected only in K562 cells, and not in Kasumi-1, MOLT-3, NIH 3T3 (Fig 1A, lanes G), and U937 (not shown) cells. K562 (Fig 1A) and MEL (not shown) were the only cell lines to express GATA-1 transcripts at detectable levels. Subgenomic, SV40 promoter-driven transcripts were present in all samples (Fig 1A). Transcripts from the GATA-modified LTR were accumulated in K562 cells at levels of approximately 50% of those derived from the wild-type LTR, as quantitated by phosphorimaging after normalization for GAPDH mRNA contents (Fig 1A, lanes G v S). The significant difference in size between the LTR-driven transcripts of GATA-SFCM and SFCM vectors is due to the use of a full-length (1.5 kb) rather than a truncated (0.9 kb) version of the LNGFR cDNA in the SFCM vector used in this particular experiment.
Cell surface expression of ΔLNGFR was quantitatively analyzed by flow cytometry in all G418-selected cell lines. Expression from the GATA-SFCM vector was observed only in K562 and MEL cells (Fig 2, dotted lines), with mean fluorescence values approximately sixfold and threefold lower, respectively (169 v 1,001 and 50 v 135 arbitrary units [AU]), than those of SFCM-transduced cells (Fig 2, bold lines). In all the other cases, ΔLNGFR expression profiles of GATA-SFCM–transduced and ΔSFCM-transduced cells were indistinguishable (NIH3T3 and Kasumi-1 are shown in Fig 2 as an example).
FACS analysis of surface ▵LNGFR expression in erythroblastic (K562 and MEL) and nonerythroblastic (3T3 and Kasumi-1) cell lines transduced by ▵SFCM (solid lines), GATA-SFCM (dotted lines), and SFCM (bold lines) vectors. Cells were stained with FITC-conjugated anti-LNGFR antibody.
FACS analysis of surface ▵LNGFR expression in erythroblastic (K562 and MEL) and nonerythroblastic (3T3 and Kasumi-1) cell lines transduced by ▵SFCM (solid lines), GATA-SFCM (dotted lines), and SFCM (bold lines) vectors. Cells were stained with FITC-conjugated anti-LNGFR antibody.
Expression of the GATA-SFCM vector is restricted to the erythroblastic progeny of transduced human CD34+ cells.
Human CD34+ cells from cord blood were transduced by multiple cycles of infection with viral supernatants in the presence of cytokines (huIL-3, huIL-6, and huSCF) and in coculture with the murine stromal cell clone CBR-BM, D#1. This clone was previously described to support survival and proliferation of human CD34+/CD38low hematopoietic stem cells,25 leading to higher yield and increased gene transfer efficiency compared with standard protocols based on cytokines only (Aiuti et al, manuscript in preparation). SFCM-transduced and GATA-SFCM–transduced CD34+ cells were analyzed for expression of ΔLNGFR by multiparameter flow cytometry 48 hours after infection. The efficiency of transduction of primitive hematopoietic progenitors, identified by a low side scatter profile and the CD34+/CD38low phenotype (Fig 3, upper panel), ranged from 55% (SFCM-transduced cells) to 23% (GATA-SFCM–transduced cells; Fig 3, lower panel), correlating with the difference in viral titer. Both wild-type and the enhancer-replaced LTRs were expressed in CD34+/CD38low cells, although at different levels (mean fluorescence of 1,003 and 214 AU, respectively). These data indicate efficient transduction of primitive hematopoietic progenitors, which were induced to proliferate without losing the primitive, CD34+/CD38low phenotype.
Multiparametric FACS analysis of human cord blood CD34+ cells 48 hours after transduction with SFCM and GATA-SFCM vectors. The forward (FSC) and side (SSC) scatter plot is in the upper-left panel. Expression of ▵LNGFR (lower panels) was analyzed in the low-SSC, CD34+, CD38low gated cell population (boxed in the upper-right panel). The percentage of ▵LNGFR+ cells (boxed) is indicated in the lower panels. Cells were stained with FITC-conjugated anti-LNGFR, TC-conjugated anti-CD38 (X axis) and PE-conjugated anti-CD34 (Y axis) antibodies. C, control, untransduced cells.
Multiparametric FACS analysis of human cord blood CD34+ cells 48 hours after transduction with SFCM and GATA-SFCM vectors. The forward (FSC) and side (SSC) scatter plot is in the upper-left panel. Expression of ▵LNGFR (lower panels) was analyzed in the low-SSC, CD34+, CD38low gated cell population (boxed in the upper-right panel). The percentage of ▵LNGFR+ cells (boxed) is indicated in the lower panels. Cells were stained with FITC-conjugated anti-LNGFR, TC-conjugated anti-CD38 (X axis) and PE-conjugated anti-CD34 (Y axis) antibodies. C, control, untransduced cells.
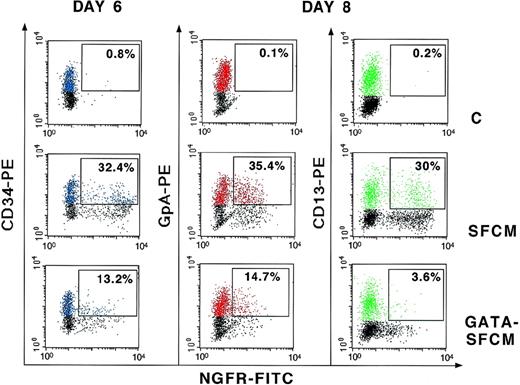
To test the expression of the chimeric LTR in the differentiated progeny of human hematopoietic progenitors, transduced CD34+ cells were grown in liquid culture in conditions that allow both myeloid and erythroid differentiation. ΔLNGFR expression was evaluated by FACS analysis of cells double-stained with antibodies against either erythroid (GpA) or myeloid (CD13) lineage-specific surface markers. A representative experiment is shown in Fig 4. After 6 days of culture and 48 hours after transduction (day 6, left panel in Fig 4), the proportion of CD34+ cells expressing the ΔLNGFR marker ranged from 32% in cultures infected with SFCM to 13% in cultures infected with GATA-SFCM, with mean fluorescence values of 1,613 and 555 AU, respectively. After induction of differentiation, ΔLNGFR was expressed in a comparable fraction of GpA+ and CD13+ cells derived from SFCM-transduced progenitors (35.4% v 30%), whereas it was expressed preferentially in GpA+ and only in a few CD13+ cells (14.7%v 3.6%) derived from GATA-SFCM–transduced progenitors (day 8, middle and right panels in Fig 4). Quantitative expression of ΔLNGFR was comparable in GpA+ cells transduced with SFCM and GATA-SFCM (246 v 218 AU), whereas in CD13+ cells transduced with GATA-SFCM, it was fivefold lower than in those transduced with SFCM (155 v 730 AU).
Expression of ▵LNGFR in liquid cultures of control (C), SFCM-transduced, and GATA-SFCM–transduced CD34+ cells. Cultures were analyzed 48 hours after transduction for expression of ▵LNGFR and CD34 (day 6) and 2 days after induction of differentiation for expression of ▵LNGFR and the lineage-specific markers GpA and CD13 (day 8). CD34+, GpA+, and CD13+ cells are shown in blue, red, and green colors, respectively. Values indicate the percentage of ▵LNGFR+cells in the CD34+, GpA+, or CD13+ fractions (boxed). Cells were stained with FITC-conjugated anti-LNGFR (X axis) and PE-conjugated anti-CD34, GpA, and CD13 (Y axis) antibodies.
Expression of ▵LNGFR in liquid cultures of control (C), SFCM-transduced, and GATA-SFCM–transduced CD34+ cells. Cultures were analyzed 48 hours after transduction for expression of ▵LNGFR and CD34 (day 6) and 2 days after induction of differentiation for expression of ▵LNGFR and the lineage-specific markers GpA and CD13 (day 8). CD34+, GpA+, and CD13+ cells are shown in blue, red, and green colors, respectively. Values indicate the percentage of ▵LNGFR+cells in the CD34+, GpA+, or CD13+ fractions (boxed). Cells were stained with FITC-conjugated anti-LNGFR (X axis) and PE-conjugated anti-CD34, GpA, and CD13 (Y axis) antibodies.
To provide an independent assay of the expression properties of the enhancer-replaced LTR, transduced CD34+ cells were grown in clonal culture in the presence of G418. Colonies were morphologically scored as BFU-E, CFU-GM, or CFU-GEMM between 10 and 14 days from plating. Transduction efficiency in clonogenic progenitors (percentage of G418-resistant colonies) ranged from 19% to 26% for the SFCM vector and from 7.5% to 12% for the GATA-SFCM vector and was overall higher for CFU-GM (15% to 51%) than for BFU-E (3.5% to 12.5%) progenitors, with no significant difference between the two vectors (data not shown). BFU-E and CFU-GM colonies were individually picked, pooled (60 to 80 colonies/pool), and analyzed for ΔLNGFR expression by immunocytochemistry. As shown in Fig 5, both BFU-E and CFU-GM colonies derived from SFCM-transduced clonogenic progenitors expressed cytoplasmic and surface ΔLNGFR (upper panels). Conversely, only BFU-E colonies derived from GATA-SFCM–transduced progenitors were positive for ΔLNGFR staining (Fig 5, lower panels). Colony identification was confirmed by staining for GpA and CD13 (not shown).
Immunocytochemical staining for ▵LNGFR in pools of BFU-E (left) and CFU-GM (right) colonies from methylcellulose clonal cultures of CD34+ hematopoietic progenitors transduced with the SFCM (upper panels) and GATA-SFCM (lower panels) vectors. ▵LNGFR+ cells appear in red after APAAP staining. The bar is 10 μm.
Immunocytochemical staining for ▵LNGFR in pools of BFU-E (left) and CFU-GM (right) colonies from methylcellulose clonal cultures of CD34+ hematopoietic progenitors transduced with the SFCM (upper panels) and GATA-SFCM (lower panels) vectors. ▵LNGFR+ cells appear in red after APAAP staining. The bar is 10 μm.
To study gene expression at the level of single colonies, CD34+ cells were transduced with the LGSN and GATA-LGSN vectors carrying the EGFP marker gene. Clonogenic assay was performed under the same conditions described for ΔLNGFR vectors, and G418-resistant, EGFP+ colonies were scored in situ under an inverted fluorescence microscope. In a representative experiment, EGFP was expressed in both CFU-GM (99 of 102) and BFU-E (48 of 50) colonies differentiated from LGSN-transduced progenitors and in most of the accessory cells in the plates (Fig 6A and B). On the contrary, EGFP expression was detected only in BFU-E colonies (13 of 14 v 1 of 30 CFU-GM colonies) in the progeny of GATA-LGSN–transduced CD34+ cells (Fig 6C through F).
Expression of EGFP in G418-resistant colonies grown in methylcellulose from CD34+ hematopoietic progenitors transduced with LGSN (A and B) and GATA-LGSN (C through F) vectors. Green-fluorescence and bright-field views of the same fields are shown in the left and right panels, respectively.
Expression of EGFP in G418-resistant colonies grown in methylcellulose from CD34+ hematopoietic progenitors transduced with LGSN (A and B) and GATA-LGSN (C through F) vectors. Green-fluorescence and bright-field views of the same fields are shown in the left and right panels, respectively.
Overall, these results show that the GATA-SFCM LTR is expressed in CD34+ hematopoietic progenitors, strongly downregulated in cells undergoing myelo-monocytic differentiation, and persistently expressed, at levels comparable to those of a wild-type LTR, in cells undergoing erythroid differentiation.
Lineage restriction of ΔLNGFR expression in mice engrafted with GATA-SFCM–transduced BM cells.
To test the expression of the enhancer-replaced LTR in the progeny of hematopoietic stem cells in vivo, 1 to 1.5 × 107 BM cells from C57Bl/6-Ly-5.1 (CD45.1+) donor mice were transduced by coculture with ecotropic SFCM and GATA-SFCM packaging cells and transplanted into lethally irradiated C57Bl/6 (CD45.2+) recipient mice. Transduction efficiency of murine progenitors, expressed as a percentage of G418-resistant colonies in methylcellulose assay, averaged 80% for SFCM and 23% for GATA-SFCM vectors, respectively. Eight weeks posttransplant, BM cells from recipient animals were analyzed for expression of ΔLNGFR and erythroid (TER-119) and myeloid (Gr-1) lineage-specific markers. Engraftment of donor cells in the BM, indicated by the proportion of CD45.1+/total CD45+ cells, averaged 66.5% ± 9.1%, with no significant difference between the two groups of mice (data not shown). In mice transplanted with SFCM-transduced BM, the proportion of ΔLNGFR+ cells averaged 3% in the Sca-1+ stem cells, 6% in the TER-119+ cells, and 4% in the Gr-1+ cells (Fig7, right panels). In recipients of GATA-SFCM–transduced BM, ΔLNGFR+ cells were detected at significant level (7%) only in the erythroblastic, TER119+ cell population (Fig 7, left panels). Quantitative expression of ΔLNGFR was comparable in TER119+ cells transduced with SFCM and GATA-SFCM (69v 116 AU). These results indicate that expression of the GATA-1–modified LTR is restricted to the erythroblastic lineage also in the progeny of transduced mouse-repopulating stem cells.
Expression of ▵LNGFR in the BM of a mouse 2 months after transplantation of SFCM-transduced (right) and GATA-SFCM–transduced (left) BM cells. Cells were stained with FITC-conjugated anti-▵LNGFR (X axis) and PE-conjugated anti-Sca1, TER 119, and Gr-1 (Y axis) antibodies. Positivity to Sca1 and TER 119 was analyzed in the blast/lymphocyte-gated cells, and positivity to GR-1 was analyzed in the granulocyte-gated cells.
Expression of ▵LNGFR in the BM of a mouse 2 months after transplantation of SFCM-transduced (right) and GATA-SFCM–transduced (left) BM cells. Cells were stained with FITC-conjugated anti-▵LNGFR (X axis) and PE-conjugated anti-Sca1, TER 119, and Gr-1 (Y axis) antibodies. Positivity to Sca1 and TER 119 was analyzed in the blast/lymphocyte-gated cells, and positivity to GR-1 was analyzed in the granulocyte-gated cells.
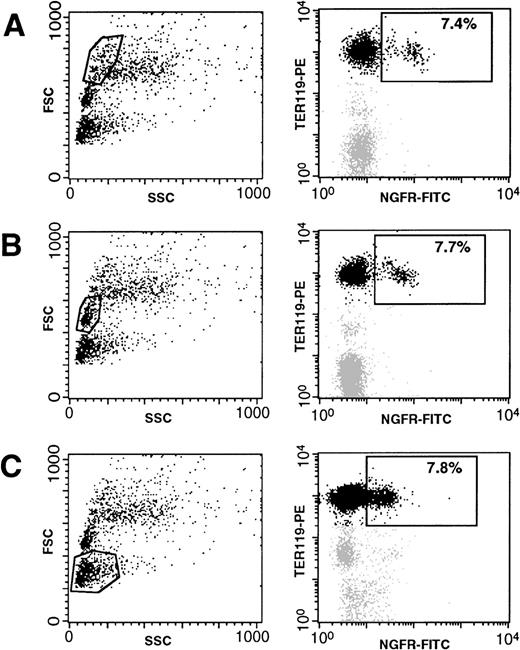
To analyze the activity of the GATA-1–modified LTR at all stages of erythroblast differentiation, expression of TER119 and ΔLNGFR was assayed in three different subpopulations of BM cells, gated according to their forward-scatter (FSC) profile. These populations contain erythroid cells at different, morphologically recognizable stages of maturation.27 As shown in Fig8, immature large erythroblasts (fraction A), more mature small erythroblasts (fraction B), and mature erythrocytes (fraction C) contain a comparable percentage of cells expressing ΔLNGFR (between 7% and 8%). However, quantitative levels of ΔLNGFR appeared to decrease with the maturation stage (mean fluorescence decreasing from 86 AU in fraction A to 22 AU in fraction C; see Fig 8). These results indicate that the GATA-1–responsive LTR is active at all stages of murine erythroid differentiation.
Expression of ▵LNGFR in erythoid cells (TER119+) at different stages of differentiation, obtained from the BM of a mouse 2 months after transplantation of GATA-SFCM–transduced BM cells. Cells were stained with FITC-conjugated anti-▵LNGFR (X axis on the right panels) and PE-conjugated anti-TER 119 (Y axis on the right panels) antibodies. Positivity to both markers was analyzed in three separate BM fractions, gated according to their forward scatter (SSC, Y axis in the left panels) profile and containing, respectively, large erythroblasts (A), mature erythroblasts (B), and erythrocytes (C).
Expression of ▵LNGFR in erythoid cells (TER119+) at different stages of differentiation, obtained from the BM of a mouse 2 months after transplantation of GATA-SFCM–transduced BM cells. Cells were stained with FITC-conjugated anti-▵LNGFR (X axis on the right panels) and PE-conjugated anti-TER 119 (Y axis on the right panels) antibodies. Positivity to both markers was analyzed in three separate BM fractions, gated according to their forward scatter (SSC, Y axis in the left panels) profile and containing, respectively, large erythroblasts (A), mature erythroblasts (B), and erythrocytes (C).
DISCUSSION
Transcriptional control of an integrated retroviral vector is mastered by the 5′ LTR of the provirus, which exerts a strong influence on the activity of any additional enhancer/promoter element placed within the viral transcription unit. Traditional ways to overcome this transcriptional interference include disabling the LTR by enhancer deletion or cloning alternative transcription units upstream of the 5′ LTR U3 region or in the opposite transcriptional orientation. Each design has advantages and limitations, and none can be considered of general applicability.6 We have tried to redirect, rather than disable, the activity of the 5′ LTR promoter by replacing the constitutive U3 extended enhancer (direct repeats + CAAT box) with the 200-bp ARE from the murine GATA-1 gene. Retroviral vectors in which the enhancer-replaced LTR drive the expression of appropriate reporter genes (ΔLNGFR or EGFP) were used to transduce human primitive hematopoietic progenitors (CD34+/CD38low) and mouse-repopulating stem cells. Analysis of transgene expression in vitro as well as in vivo indicated that the GATA-1 ARE effectively restricts transcription from the LTR promoter to the erythroid progeny of the transduced stem cells. The activity of the modified LTR was in fact quantitatively comparable to that of the wild-type LTR in erythroblasts and erythropoietic cell lines, at the level of both RNA accumulation and protein synthesis, whereas it was significantly lower in human CD34+/CD38low and murine Sca1+progenitors and virtually absent in other cell types. Transcription of the modified LTR appears to be activated in cycling progenitors, downregulated in myelo-monocytic cells, and maintained at sustained levels in all stages of erythroid differentiation, strictly paralleling the expression pattern of the GATA-1 gene during differentiation of human primary hematopoietic cells.28 29 The LTR modification had no effect on integration and stability of the provirus in the target cell genome. Enhancer replacement is therefore an effective strategy to target transcription of a retroviral genome to a specific progeny of stem cells transduced ex vivo and reintroduced into an active hematopoietic system.
These data provide new insight into the role of the ARE in the physiological regulation of the GATA-1 gene. In adult mice, expression of GATA-1 is restricted to hematopoietic cells and Sertoli cells of the testis by the activity of two alternative promoters, designated IE and IT, located approximately 8 kb apart on the X chromosome.30The proximal, erythroid-specific IE promoter drives transcription of GATA-1 at low levels in multipotential hematopoietic progenitors and at high levels in differentiating erythroblasts, megacaryocytes, and mast cells.31 Initiation and maintenance of GATA-1 transcription in development and differentiation of both primitive and definitive erythropoietic cells was shown to be under the control of multiple, DNase hypersensitive elements located upstream (from −3,900 to −2,700) and downstream (first intron) of the IE promoter.32-35 The ARE (−856 to −655 from the IE promoter), which contains two GATA-1 binding sites, was originally identified by in vitro footprinting17 and shown to restrict expression of a heterologous promoter to erythroblastic cell lines in a transient transfection assay.17,18 Although its role has not been addressed by direct mutagenesis in vivo and remains therefore uncertain, this element might cooperate in erythroid-specific maintenance of GATA-1 gene expression by activating a GATA-1–driven auto-regulatory loop.17,18 Our data provide the first in vivo evidence for such a role by showing that transcription of a transgene under the control of the GATA-1 ARE is activated in early hematopoietic progenitors (Sca1+) and maintained at significant levels throughout erythropoietic differentiation with a kinetics virtually overlapping that of the endogenous GATA-1 gene. The auto-regulatory circuit established by the ARE is therefore sufficient to maintain GATA-1–dependent transcription in differentiating erythroblasts. This circuit might be essential to sustain GATA-1 transcription at appropriate levels in erythroid cells, the maturation of which is dose-dependent with respect to GATA-1 levels.33
As a strategy for transcriptional targeting, manipulating the LTR control elements offers a number of advantages. First, it provides a more efficient vector design, which allows us to use the major viral transcription unit to express the gene of interest under the form of a spliceable RNA. Second, it allows the use of a second, independently regulated internal promoter to control an additional function, such as a positive (NeoR or ΔLNGFR) or negative (HSV-TK) selectable marker for selecting packaging cells or eliminating transduced cells in vivo. Third, the presence of a genomic control element into the LTR could reduce the chances of chromatin-mediated inactivation of the integrated provirus. This is known to affect the long-term maintenance of MoMLV LTR-driven gene expression particularly in stem cells36-38and has been circumvented in the past by replacing the MoMLV enhancer with that of viruses with different tropism.39-41 The use of genomic instead of viral sequences could have the additional advantage of promoting transcriptionally active chromatin configurations only in the cells in which the transgene is transcriptionally targeted.
We had previously shown that the simple insertion of a muscle-specific enhancer into an intact MoMLV LTR is sufficient to restrict the transcriptional activity of the viral promoter to differentiated muscle fibers.13 However, the same design was unsuccessful in the case of the GATA-1 ARE, which has no effect in modulating the activity of the LTR if inserted upstream of the viral enhancer (G.F., unpublished results). Replacing, rather than adding sequences to, the viral enhancer therefore appears to be a more efficient strategy to restrict transcriptional activity of retroviral vectors, although its general applicability needs to be tested in additional contexts. In fact, the LTR architecture imposes a number of constrains upon the elements that it can accommodate, such as the distance between the enhancer and the promoter and the sequence of the enhancer itself, and it is reasonable to assume that some tissue-or cell-specific elements could be unable to drive the TATA-containing LTR promoter in an appropriate fashion. Unpredictable behavior of chimeric LTRs has already been observed in the past.42 Maintaining the LTR CAAT element downstream from the replaced enhancer increases the transcriptional efficiency of the chimeric LTR by increasing the activity of the basal promoter, although this necessarily limits the stringency of the tissue- or cell lineage-specificity14(see Results). The overall size of the inserted fragment can also be a problem: an LTR much bigger than its normal size reduces both viral titer and stability of proviral integration, most likely because of decreased efficiency of reverse transcription.14,43 44Although titer problems could be overcome in a number of ways, a high frequency of rearrangement would limit the practical use of certain enhancer-replaced vectors.
The vector targeting strategy described in this paper could be potentially relevant for gene therapy of hemoglobinopathies. Current attempts to build retroviral vectors for expression of β-globin in erythropoietic cells are based on the use of a full β-globin gene and part of the β-globin locus control region (LCR), with the rationale of reproducing, on a smaller size scale, the high-level and position-independent expression provided by the interaction between the LCR and the β-globin promoter.45,46 However, genetic instability severely limits the possibility of using full-length LCR sequences in the context of a retroviral vector,47,48whereas small LCR fragments are apparently unable to reduce the integration-dependent variability of transgene expression observed with MoMLV-based vectors.24,49 Indeed, the very concept that the LCR has a role in maintaining an open chromatin structure to the globin locus has been challenged by more recent evidence.50,51Binding of erythroid-specific transcription factors, such as GATA-1, to enhancers of erythroid-specific genes early in development or differentiation could instead be a key factor in initiation and maintenance of open chromatin configurations.52,53 Although our experiments do not directly address this issue, LTRs engineered with GATA-1–responsive elements could therefore provide a certain degree of position-independence to the expression of integrated proviruses in erythroid cells. A potential limitation of such a vector design could be an insufficient level of β-globin mRNA accumulation, which even in the best conditions (use of a β-globin promoter and noncoding trailer sequences), is likely to remain lower than that obtainable with an LCR. However, even a mild correction of the β-globin imbalance in maturing erythroblasts might be sufficient to reduce ineffective erythropoiesis in severe β-thalassemia patients54 and significantly affect both morbidity and clinical management of the disease. On the short-term, a limited-correction gene therapy strategy based on autologous transplantation of BM transduced with a transcriptionally targeted retroviral vector could therefore be a safe and viable alternative to a still elusive full functional replacement of the β-globin locus.
ACKNOWLEDGMENT
The authors thank C. Moroni for the initial help in constructing and testing hybrid LTRs and G. Torriani and R. Parma for the excellent technical assistance.
Supported by a core grant from the Italian Telethon Foundation and a grant from the Istituto Superiore di Sanità.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Giuliana Ferrari, PhD, TIGET-H.S. Raffaele, Via Olgettina, 58, 20132 Milano, Italy; e-mail:ferrari@tigem.it.