Abstract
Chromosomal abnormalities in acute leukemia have led to the discovery of many genes involved in normal hematopoiesis and in malignant transformation. We have identified the fusion partners in an inv(8)(p11q13) from a patient with acute mixed lineage leukemia. We show by fluorescence in situ hybridization (FISH) analysis, Southern blotting, and reverse transcriptase-polymerase chain reaction (RT-PCR) that the genes for MOZ, monocytic leukemiazinc finger protein, and TIF2,transcriptional intermediary factor 2, are involved in the inv(8)(p11q13). We demonstrate that the inversion creates a fusion between the 5′ end of MOZ mRNA and the 3′ end of TIF2 mRNA maintaining the translational frame of the protein. The predicted fusion protein contains the zinc finger domains, the nuclear localization domains, the histone acetyltransferase (HAT) domain, and a portion of the acidic domain ofMOZ, coupled to the CREB-binding protein (CBP) interaction domain and the activation domains of TIF2. The breakpoint is distinct from the breakpoint in the t(8;16)(p11;p13) translocation in acute monocytic leukemia with erythrophagocytosis that fuses MOZ with CBP. The reciprocalTIF2-MOZ fusion gene is not expressed, perhaps as a result of a deletion near the chromosome 8 centromere. TheMOZ-TIF2 fusion is one of a new family of chromosomal rearrangements that associate HAT activity, transcriptional coactivation, and acute leukemia.
© 1998 by The American Society of Hematology.
SPECIFIC RECURRING chromosomal abnormalities are commonly associated with acute myeloid leukemia.1 These chromosomal anomalies influence the molecular and cellular phenotype of the leukemic blasts and may be responsible for their malignant potential.2 The aberrations often lead to the formation of one or more fusion genes resulting in the overexpression or untimely expression of a normal gene, eg, theMYC/Ig gene enhancer fusion produced by the t(8;14) in Burkitt’s lymphoma,3,4 or the creation of a new gene product by fusing genes as in the PML-RAR fusion produced by the t(15;17) characteristic of acute promyelocytic leukemia.5 6 Some regions are common partners in fusion events; 11q23 is involved in at least 40 different translocations in acute leukemia.
Two distinct clinical syndromes have been associated with 8p11: a chronic myeloproliferative disorder complicated by T-cell lymphoblastic leukemia/lymphoma and peripheral blood eosinophilia and a M4/M5 acute monocytic leukemia with prominent erythrophagocytosis.7-11The t(8;16) translocation in acute monocytic leukemia with erythrophagocytosis fuses the gene for MOZ with the transcriptional coactivator CBP.10 The t(8;13) translocation associated with a chronic myeloproliferative disorder, lymphoblastic lymphoma, and eosinophilia juxtaposes the gene forFGFR1 with a novel gene that encodes the zinc finger protein,ZNF198.12 These translocations appear to involve different portions of 8p11 than the t(8;22) translocation observed in acute monocytic leukemia.8
We have identified the genes involved in a novel fusion event in a case of acute mixed-lineage leukemia associated with an inv(8)(p11q13). The patient’s clinical presentation is distinct from the myeloproliferative/lymphoblastic disorder and the M4/M5 leukemia with erythrophagocytosis previously associated with 8p11 abnormalities, and the fusion event creates a chimeric gene from the potential histone transacetylase, MOZ, and the transcriptional coactivator,TIF2.
MATERIALS AND METHODS
Patient.
A 29-year-old female presented after 1 month of back pain, 2 weeks of extensive bruising, and a few hours of rectal bleeding. She had 102,000 blasts/μL that were French-American-British (FAB) M0/M1 by morphology; M7 by histochemical staining; positive for CD5, CD10, HLA-DR, and CD33; and negative for CD34, CD19, CD13, and CD7. Cytogenetic analysis of bone marrow cells showed that 3 cells were 46, XX and that 17 cells were inv(8)(p11q13), der(10) t(1;10)(q25;p15) (Fig 1A). The patient was enrolled and treated on SWOG study 9034, but relapsed 30 days after induction. Remission was attained after allogeneic bone marrow transplantation, but 67 days after transplantation she presented with headaches. Stereotactic biopsy of her brain lesion showed a chloroma and, despite cranial irradiation, she suffered intraparenchymal cerebellar hemorrhage and died.
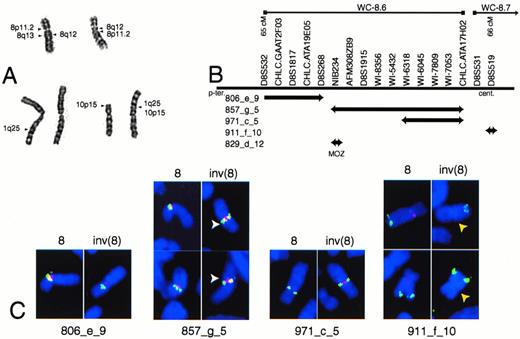
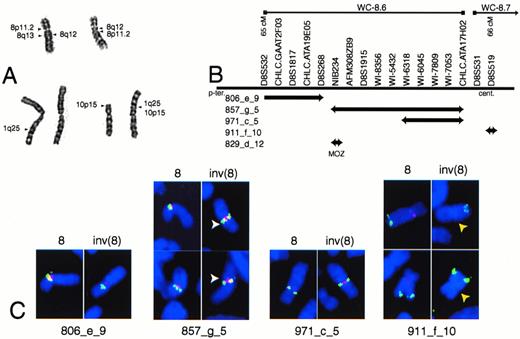
EBV-LEUK1 cytogenetics and FISH analyses. (A) A partial karyotype of G-banded chromosomes 1, 8, and 10 from EBV-LEUK1 cells. The arrows indicate regions of chromosomal fusion. (B) Map of the YACs versus genetic markers from singly linked contigs WC-8.6 and WC-8.7. The relative size of the YACs and the spacing of the markers are not drawn to scale and are not precisely known. (C) Examples of FISH performed on EBV-LEUK1 cells with YAC probes from the human CEPH library. YACs 806_e_9, 857_g_5, and 971_c_5 appear yellow-green. The chromosome 8 centromeric probe appears red. YAC 911_f_10 and the 8q24 probe (MYC) appear red and yellow, respectively. DAPI-stained chromosomes are blue. White arrows highlight the split signal obtained with YAC 857_g_5, and yellow arrows highlight the lack of binding to 911_f_10.
EBV-LEUK1 cytogenetics and FISH analyses. (A) A partial karyotype of G-banded chromosomes 1, 8, and 10 from EBV-LEUK1 cells. The arrows indicate regions of chromosomal fusion. (B) Map of the YACs versus genetic markers from singly linked contigs WC-8.6 and WC-8.7. The relative size of the YACs and the spacing of the markers are not drawn to scale and are not precisely known. (C) Examples of FISH performed on EBV-LEUK1 cells with YAC probes from the human CEPH library. YACs 806_e_9, 857_g_5, and 971_c_5 appear yellow-green. The chromosome 8 centromeric probe appears red. YAC 911_f_10 and the 8q24 probe (MYC) appear red and yellow, respectively. DAPI-stained chromosomes are blue. White arrows highlight the split signal obtained with YAC 857_g_5, and yellow arrows highlight the lack of binding to 911_f_10.
Epstein-Barr virus (EBV)-transformed cell line.
Frozen primary leukemic blasts (LEUK1) were infected with EBV and grown without cyclosporin A. Two months after infection, one culture (EBV-LEUK1) began active proliferation. Cytogenetic analysis of EBV-LEUK1 showed an inv(8)(p11q13) and a der(10)t(1;10)(q25;p15) in all metaphases. Flow cytometric analysis of EBV-LEUK1 showed loss of the CD10 and gain of CD7. The cells were also positive for CD33, CD13, and CD41.
Fluorescence in situ hybridization (FISH).
EBV-LEUK1 metaphase spreads were prepared after cell cycle synchronization, inhibition of chromosome condensation, and metaphase arrest with colcemid. Total yeast DNA was isolated by the spheroplast method from yeast artificial chromosome (YAC) clones from the human CEPH library (Research Genetics, Inc, Huntsville, AL). Human DNA was labeled with biotin or digoxigenin (1:3 ratio to dT; Boehringer Mannheim, Indianapolis, IN) byAlu-polymerase chain reaction (PCR) with Alu primers CL1 and CL2 (Table 1). Probes were combined with human cot-1 DNA in Hybrisol VII (Oncor, Gaithersburg, MD) before hybridization and detected with Cy3-streptavidin (Jackson ImmunoResearch, West Grove, PA), fluorescein isothiocyanate (FITC)-avidin (Oncor), FITC-antidigoxigenin, or rhodamine-antidigoxigenin (Oncor). Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Southern blot analysis.
DNA from LEUK1 cells and normal peripheral blood mononuclear cells was digested with restriction enzymes, size-fractionated, and transferred to Nytran Plus membranes (Schleicher & Schuell, Keene, NH). A MOZ probe generated by PCR (MOZ2549F to 3663R) was labeled with α-32P-dCTP by random priming. After hybridization and stringent washing, the blot was exposed to a phosphorimager screen.
Reverse transcriptase-PCR (RT-PCR).
EBV-LEUK1 cells were lysed with guanidinium thiocyanate. After homogenization, total cellular RNA was collected through 5.7 mol/L CsCl. Total cellular RNA (0.5 μg) was denatured at 75°C for 5 minutes with 50 pmol of random hexamers. First-strand cDNA was prepared in a 20 μL reaction containing 2.5 U of Moloney’s murine leukemia virus (MMLV) reverse transcriptase (Superscript RT; GIBCO BRL, Gaithersburg, MD), 20 mmol/L Tris-HCl, pH 7.5, 50 mmol/L KCl, 10 mmol/L MgCl2, 2.5 mmol/L dithiothreitol, 0.5 U RNasin (Promega, Madison, WI), and 625 μmol/L of each dNTP. Twenty-microliter PCR reactions, containing 5 pmol each of MOZ and/or TIF2 primers, 0.2 mmol/L dNTPs, 1× Vent buffer, 0.5 U Taq (Fisher, Pittsburgh, PA), and 0.2 U Vent (New England Biolabs, Beverly, MA) DNA polymerase, were incubated at 94°C for 10 seconds, 55°C for 30 seconds, and 72°C for 4 minutes for 10 cycles, followed by 25 cycles with a 20-second increase in elongation time/cycle.
RESULTS
FISH.
A map showing the relative positions of YACs is presented (Fig 1B). YAC probes from contigs WC-8.5, WC-8.6, and WC-8.7 (Whitehead Institute, Cambridge, MA) were hybridized to chromosome spreads from EBV-LEUK1. Chromosome 8 was identified by hybridization with a chromosome 8 centromere probe or a probe containing MYC(YAC 934_e_1, band 8q24). YAC 806_e_9 from contig WC-8.6 is telomeric to the breakpoint on the p-arm and appears more telomeric on the inv(8) chromosome than on the normal chromosome 8. YAC 857_g_5 hybridizes as a split signal on the inv(8) chromosome (Fig 1C). The telomeric end of YAC 857_g_5 completely overlaps YAC 829_d_12, which contains the gene for MOZ.9 10 YAC 971_c_5 is adjacent to the centromere on 8p, but becomes pericentrometic after the paracentric inversion. YAC 911_f_10 from contig WC-8.7 is pericentromeric on the wild-type chromosome, but absent from the inv(8) chromosome. The lack of a hybridization signal for YAC 911_f_10 suggests the inv(8) contains a deletion near the centromere (Fig 1C).
Southern blot analysis for MOZ.
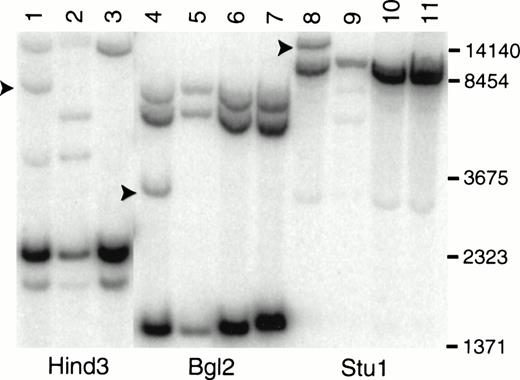
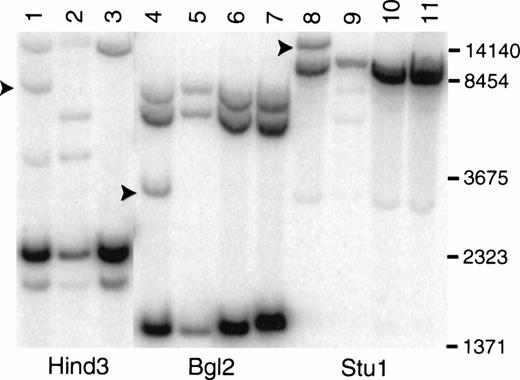
The involvement of MOZ in the breakpoint at 8p11 is supported by Southern blot analysis of DNA from primary leukemic cells (Fig 2). A MOZ cDNA probe detects rearranged fragments of 3.5, 8.0, and 15 kb after digestion with Bgl 2, Hind 3, and Stu 1 (Fig 2, lanes 4, 1, and 8), respectively. Hind 3 and Stu 1 restriction fragment length polymorphisms were identified in DNA from normal control no. 1 (Fig 2, lanes 2 and 9).
Southern blot analysis of LEUK1 and normal control DNA probed with a MOZ cDNA probe. High molecular weight DNA was digested to completion with Hind 3, Bgl 2, or Stu 1; size-fractionated; blotted; and hybridized to a probe that extends from bp 2549 to 3663 in the MOZ cDNA. The blot was washed in 0.1× SSC at 65°C for 30 minutes. Lanes 1, 4, and 8 contain DNA from LEUK1 cells; lanes 2, 5, and 9 contain DNA from normal volunteer no. 1; lanes 3, 6, and 10 contain DNA from normal volunteer no. 2; and lanes 7 and 11 contain DNA from normal volunteer no. 3. The arrows indicate the positions of the rearranged restriction fragments in LEUK1 DNA. Normal volunteer no. 1 has restriction fragment length polymorphisms after Hind 3 and Stu 1 digestion (lanes 2 and 9).
Southern blot analysis of LEUK1 and normal control DNA probed with a MOZ cDNA probe. High molecular weight DNA was digested to completion with Hind 3, Bgl 2, or Stu 1; size-fractionated; blotted; and hybridized to a probe that extends from bp 2549 to 3663 in the MOZ cDNA. The blot was washed in 0.1× SSC at 65°C for 30 minutes. Lanes 1, 4, and 8 contain DNA from LEUK1 cells; lanes 2, 5, and 9 contain DNA from normal volunteer no. 1; lanes 3, 6, and 10 contain DNA from normal volunteer no. 2; and lanes 7 and 11 contain DNA from normal volunteer no. 3. The arrows indicate the positions of the rearranged restriction fragments in LEUK1 DNA. Normal volunteer no. 1 has restriction fragment length polymorphisms after Hind 3 and Stu 1 digestion (lanes 2 and 9).
Identification of the fusion partner.
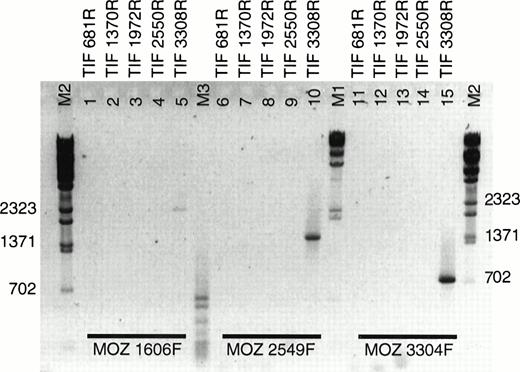
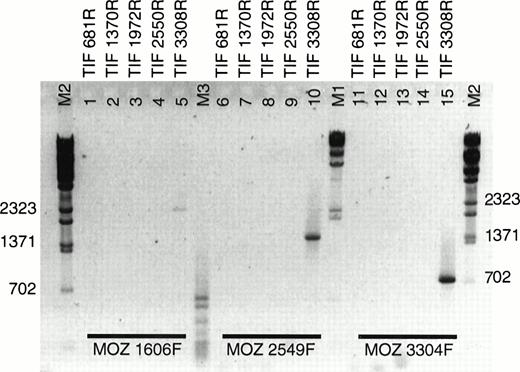
During our analysis of the MOZ fusion partner by rapid amplification of cDNA ends (RACE), a fusion ofMOZ to a gene on 8q13 was reported in a case of acute monocytic leukemia.13 In this case, MOZ was fused to the gene for TIF2. The presence of a fusion between MOZ andTIF2 in EBV-LEUK1 cells was tested by RT-PCR. Five antisense primers evenly spaced across the coding sequence of TIF2 were paired with 3 sense primers derived from MOZ (Table 1). TIF 3308R, when combined with MOZ primers 1606F, 2549F, and 3304F, supported the amplification of PCR products of approximately 2,470, 1,530, and 775 bp (Fig 3, lanes 5, 10, and 15), suggesting that the breakpoint is flanked by MOZ 3304F and TIF 3308R. None of the TIF2 primers derived from sequence 5′ of TIF 3308R resulted in successful amplification when paired withMOZ primers.
RT-PCR of LEUK1 RNA with MOZ and TIF2primers. cDNA from EBV-LEUK1 was amplified by PCR, and products were subjected to electrophoresis in 2% agarose gels in 1× TAE. Three sense primers from MOZ were paired with 5 antisense primers from TIF2. Specific PCR products were produced when eachMOZ primer was combined with TIF 3308R (lanes 5, 10, and 15). The M2 indicates λ-BstE 2 DNA size markers.
RT-PCR of LEUK1 RNA with MOZ and TIF2primers. cDNA from EBV-LEUK1 was amplified by PCR, and products were subjected to electrophoresis in 2% agarose gels in 1× TAE. Three sense primers from MOZ were paired with 5 antisense primers from TIF2. Specific PCR products were produced when eachMOZ primer was combined with TIF 3308R (lanes 5, 10, and 15). The M2 indicates λ-BstE 2 DNA size markers.
Expression of wild-type MOZ and TIF2 by RT-PCR.
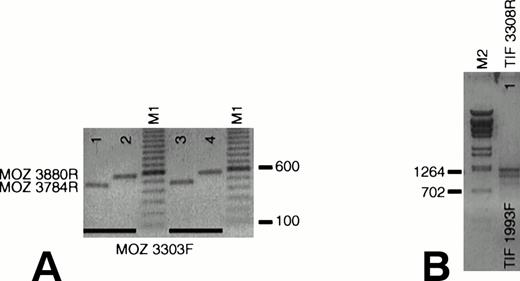
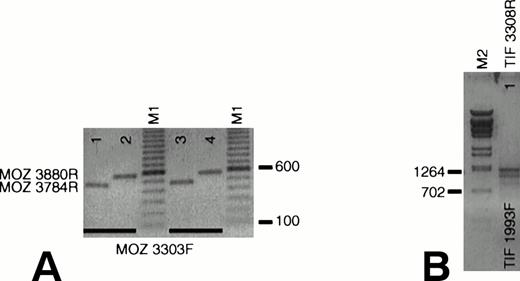
We queried whether wild-type MOZ and TIF2 are expressed in EBV-LEUK1 cells. Amplification with primers that flank theMOZ breakpoint (MOZ 3303F v MOZ 3784R and MOZ 3880R) generated PCR products of 482 and 578 bp with cDNA prepared from normal peripheral blood mononuclear (Fig 4A, lanes 1 and 2) and EBV-LEUK1 cells (Fig 4A, lanes 3 and 4). Appropriately sized products (1,316 bp/1,109 bp) were also obtained with primers that flank the TIF2 breakpoint, TIF 1993F and TIF 3308R (Fig 4B). This region of TIF2 contains a 207-bp alternatively spliced exon, giving rise to two PCR products.
Expression of wild-type MOZ and TIF2. (A) RNA from EBV-LEUK1 and normal peripheral blood mononuclear cells was analyzed by RT-PCR. A PCR product containing the MOZ breakpoint was amplified when MOZ 3303F was paired with either MOZ 3784R (lanes 1 and 3) or MOZ 3880R (lanes 2 and 4). (B) TIF2 primers that flank the TIF2 breakpoint amplify products of 1,315 and 1,105 bp from EBV-LEUK1 cDNA, corresponding to the alternatively spliced forms of TIF2 cDNA. M1 and M2 indicate 100-bp marker ladders and λ-BstE 2 DNA size markers, respectively.
Expression of wild-type MOZ and TIF2. (A) RNA from EBV-LEUK1 and normal peripheral blood mononuclear cells was analyzed by RT-PCR. A PCR product containing the MOZ breakpoint was amplified when MOZ 3303F was paired with either MOZ 3784R (lanes 1 and 3) or MOZ 3880R (lanes 2 and 4). (B) TIF2 primers that flank the TIF2 breakpoint amplify products of 1,315 and 1,105 bp from EBV-LEUK1 cDNA, corresponding to the alternatively spliced forms of TIF2 cDNA. M1 and M2 indicate 100-bp marker ladders and λ-BstE 2 DNA size markers, respectively.
Expression of the TIF2-MOZ fusion.
The chromosomal inversion that created the MOZ-TIF2fusion at 8p11 must result in a reciprocal genomic fusion at 8q13. The expression of a TIF2-MOZ fusion mRNA in EBV-LEUK1 was tested with sense primers 5′ of the TIF2 breakpoint and antisense primers 3′ of the MOZ breakpoint. No PCR products were generated with these primers, suggesting that the reciprocal fusion mRNA is not expressed (data not shown).
Identification of the breakpoint.
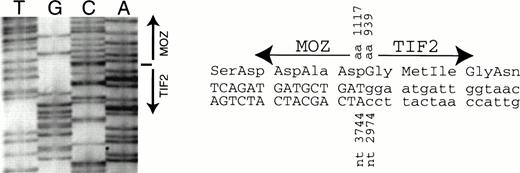
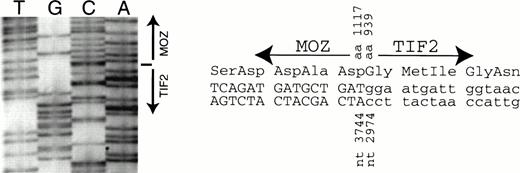
The PCR product obtained from EBV-LEUK1 cDNA with MOZ 2549F andTIF2 4939R primers was subjected to DNA sequencing. The breakpoint between the cDNAs for TIF2 and MOZ occurs at bp 2974 and 3744, respectively (Fig 5A). The breakpoint creates an in-frame fusion of MOZ andTIF2 at amino acids 1117 and 939, respectively (Fig 5B).
DNA sequence across the breakpoint. An RT-PCR product from EBV-LEUK1 RNA that extends from MOZ 2549F to TIF2 4939R was subcloned. (A) The subcloned DNA was sequenced with primer TIF2 3308R. The arrow indicates the breakpoint between MOZ and TIF2at nucleotides 2974 and 3744, respectively. (B) The DNA sequence around the breakpoint and the predicted amino acid sequence is presented.
DNA sequence across the breakpoint. An RT-PCR product from EBV-LEUK1 RNA that extends from MOZ 2549F to TIF2 4939R was subcloned. (A) The subcloned DNA was sequenced with primer TIF2 3308R. The arrow indicates the breakpoint between MOZ and TIF2at nucleotides 2974 and 3744, respectively. (B) The DNA sequence around the breakpoint and the predicted amino acid sequence is presented.
DISCUSSION
Clinical syndromes with 8p11 abnormalities include a chronic myeloproliferative disorder associated with T-cell lymphoblastic lymphoma and eosinophilia [t(8;13)(p11;q11-12), t(8;9)(p11;q34), t(6;8)(q27;p11)]7,8,11 and an acute myelomonocytic leukemia with erythrophagocytosis [t(8;16)(p11;p13), inv(8)(p11q13), t(8;22)(p11;q13), and t(8;19)(p11;q13)].10,14,15 Two genes from the 8p11 locus have been identified in these diseases:MOZ, isolated by positional cloning of the breakpoint in t(8;16),10 and FGFR1, cloned as the chromosome 8 contribution to the t(8;13).12 The 8p11 genes are fused toCBP and ZNF198, respectively. ZNF198 provides a zinc finger dimerization motif to the tyrosine kinase domain ofFGFR1, creating a fusion protein that is predominantly cytoplasmic and probably constitutively activated by homodimerization.12 Both MOZ10,16 andCBP17-19 have been associated with chromatin remodeling and implicated in histone acetylation.
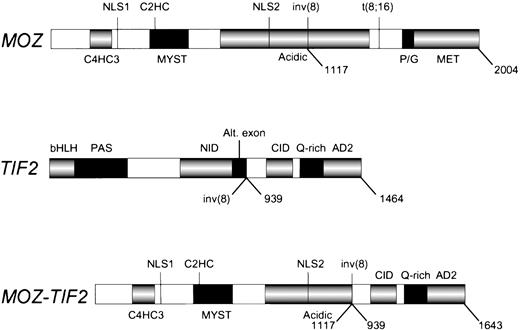
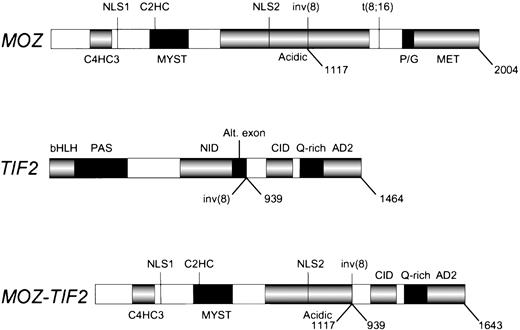
We have identified a gene fusion created by an inv(8)(p11q13) in a case of de novo acute mixed lineage leukemia. The 8p11 gene encodesMOZ, a 2,004 amino acid protein with 2 types of zinc fingers and a HAT signature motif,10 and the 8q13 gene encodesTIF2, a transcriptional coactivator with nuclear receptor binding domains, a CBP interaction domain and two activation domains.20,21 The fusion gene produces a mRNA containing the 5′ end of MOZ appended in translational frame to the 3′ end of TIF2. The breakpoints occur at nucleotide 3744 of MOZ (corresponding to amino acid 1117) and 2974 ofTIF2 (corresponding to amino acid 939). The inv(8) MOZbreakpoint is distinct from the breakpoint in the MOZ-CBPfusion in the t(8;16)(p11;p13) (nucleotide 5033, amino acid 1547).10 The fusion product retains the C4HC3 and C2HC zinc fingers, two putative nuclear localization signals, the HAT domain, the MYST domain and a portion of the acidic domain of MOZ, and theCBP interaction domain, the Q-rich region, and two activation domains of TIF2 (Fig 6). The breakpoint in TIF2 occurs at the splice acceptor site for an alternatively spliced exon,20 suggesting that the genomic breakpoint is within an intron. We have verified expression of wild-type MOZ and two known variants of TIF2 in EBV-LEUK1 cells by amplifying cDNA with primers that flank either theMOZ or the TIF2 breakpoint. The TIF2-MOZreciprocal fusion is not expressed at the sensitivity of RT-PCR, perhaps due to loss of genetic material from the promoter or N-terminal coding region of TIF2. If the genomic deletion on the inv(8) chromosome involves TIF2, the C-terminal portion of MOZmay have joined with an unknown gene adjacent to TIF2; hence, the possibility of an expressed reciprocal fusion has not been entirely eliminated.
The MOZ, TIF2, and MOZ-TIF2fusion proteins. The protein domains of MOZ, TIF2, and the MOZ-TIF2 fusion are presented. C4HC3, zinc finger domain associated with chromatin binding; C2HC, zinc finger domain associated with HAT activity; HAT, histone acetyltransferase; NLS, nuclear localization signals; MYST, MOZ, YBF2, SAS2, and Tip60 homology region; inv(8), breakpoint in acute mixed lineage leukemia; Acidic, acidic domain; t(8;16), breakpoint in AML with erythrophagocytosis; P/G, proline/glutamine-rich region; MET, methionine-rich region; bHLH, basic helix-loop-helix region; PAS,Per/ARNT/Sim homology region; Alt.exon, alternatively spliced exon (AA 869 to 938); AD2, activation domain-2; CID, CBPinteraction domain (contains activation domain-1); NID, nuclear hormone receptor interaction domain; Q-rich, glutamine-rich region. The numbers below each schematic correspond to amino acids.
The MOZ, TIF2, and MOZ-TIF2fusion proteins. The protein domains of MOZ, TIF2, and the MOZ-TIF2 fusion are presented. C4HC3, zinc finger domain associated with chromatin binding; C2HC, zinc finger domain associated with HAT activity; HAT, histone acetyltransferase; NLS, nuclear localization signals; MYST, MOZ, YBF2, SAS2, and Tip60 homology region; inv(8), breakpoint in acute mixed lineage leukemia; Acidic, acidic domain; t(8;16), breakpoint in AML with erythrophagocytosis; P/G, proline/glutamine-rich region; MET, methionine-rich region; bHLH, basic helix-loop-helix region; PAS,Per/ARNT/Sim homology region; Alt.exon, alternatively spliced exon (AA 869 to 938); AD2, activation domain-2; CID, CBPinteraction domain (contains activation domain-1); NID, nuclear hormone receptor interaction domain; Q-rich, glutamine-rich region. The numbers below each schematic correspond to amino acids.
The involvement of MOZ, CBP, and TIF2 in acute leukemia suggests that yet another class of cellular processes can be deranged by gene fusion events. Each of these proteins possesses or recruits HATs to chromatin. The ability of HATs to affect chromatin structure and regulate gene expression is well appreciated.22 How the MOZ-TIF2 fusion protein is involved in acute leukemia is not known. We speculate that the TIF2 portion of the fusion associates with CBPproviding additional activation domains for stimulating transcription or additional HAT activity for remodeling chromatin. The protein-protein dimerization faces of MOZ could deliver theCBP interaction domain and the activation domains ofTIF2 to unique chromosomal locations, modifying transcription by allowing binding of CBP or other proteins. The fusion protein could function as a dominant negative by interacting with normal MOZ, TIF2, and/or CBP on or off the surface of chromatin. We hope the discovery of theMOZ-TIF2 fusion will uncover basic relationships between the control of chromatin condensation and gene expression in acute leukemia and suggest new targets for directed therapeutic intervention.
Address reprint requests to Kerry L. Blanchard, PhD, MD, Louisiana State University Medical Center, 1501 Kings Hwy, Shreveport, LA 71103; e-mail: Kblanc@lsumc.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.