Abstract
The G1-phase cell-cycle inhibitor p21 has been proposed to mediate growth arrest during differentiation. Upregulation of p21 has been shown in multiple cell lines induced to differentiate; however, the mechanism of p21 induction during normal differentiation is largely unknown. In this report, we use normal hematopoietic precursor cells obtained from umbilical cord to model p21 regulation during differentiation. Myeloid maturation of CD34+ precursor cells is associated with a marked increase in p21 expression at the RNA and protein level. The upregulation of p21 transcripts during differentiation is associated with decreased binding to a highly conserved 44-bp fragment within the p21 promoter. This 44-bp regulatory element binds a novel modulator of p21 expression. It is of considerable interest that, although the binding activity is expressed in p53-negative as well as in p53-positive cells, the DNA sequence recognized by this protein overlaps a PuPuPuC(A/T)(T/A)GPyPyPy consensus sequence for p53.
NUMEROUS STUDIES1-4 have confirmed the association between differentiation commitment and arrest in the G1 phase of the cell cycle, but mechanisms of the linkage are incompletely understood. The cell cycle inhibitor p215-8may play a role in the growth arrest of differentiated cells. p21 inhibits the ability of G1-phase cyclin-cdk complexes to phosphorylate sustrates such as the members of the Rb gene family.5Through its antagonism of cdk action and potentially by interfering with PCNA processivity,9 high levels of p21 arrest cells in G1 phase. p21 may contribute to the growth arrest of differentiated cells by supporting the ability of Rb, p107, and/or p130 to bind and sequester E2F factors.3,10-15 Several studies have demonstrated that p21 expression is upregulated during differentiation of hematopoietic cells16-20 and may promote the transition into the differentiated state.21 Subsequent studies have confirmed the association of p21 expression and differentiation both through in vitro differentiation models17,22 and through in situ staining of p21 message in embryonic and adult tissues.23,24 Cell cycle arrest mediated by p21 may also inhibit apoptosis of differentiated cells.25-28
The mechanism of p21(WAF1) induction during differentiation is largely unknown. p21 is induced during DNA damage through the action of p53 at discrete sites within the p21 promoter.23,29 However, the p53 pathway is unlikely to modulate p21 upregulation during differentiation, because p21 and p53 are expressed in distinct patterns on immunohistochemical staining of colon, keratinocytes, and mouse embryo and because p21 induction occurs in p53-null models of differentiation.16,17,23,24,30-32 Regulation of p21 during differentiation is likely to be complex and involve transcriptional adaptor molecules such as p30031 as well as tissue-specific factors. Analysis of p21 induction in monocyte/macrophage cell line models may be particularly pertinent to hematopoietic differentiation. Induction of U937 myelomonocytic cells by phorbol ester or okadaic acid to differentiate into macrophages is associated with p21 upregulation mediated by recruitment of the Sp1 transcription factor to binding sites adjacent to the transcription startsite.20 Vitamin D3 also promotes U937 differentiation and upregulates p21, acting through a response element in the p21 promoter.33 As downstream effectors of cytokines, Stat134 and Stat 535 induction of p21 could also contribute to p21 modulation during hematopoiesis. However, modulators of p21 expression that are active in normal hematopoietic development and are stage-specific in their action have yet to be reported. Indeed, the normal pattern of p21 expression during myelomonocytic differentiation of normal cells has not previously been characterized. In this report, we present data that characterize p21 expression in CD34+ hematopoietic precursor cells as they are induced to differentiate along the myelomonocytic lineage. Additionally, a highly conserved, transcriptionally active fragment in the p21 promoter is demonstrated to bind proteins specific to distinct stages of hematopoietic maturation. This activity is distinct from previously reported modulators of p21 expression, including p53, although a p53-response element is contained within the binding sequence. The binding activity is inversely correlated with p21 expression, suggesting that a transcriptional repressor is involved. Moreover, the binding activity appears to be inhibited by protein extracts of more mature cells. A subset of leukemic cells exhibit aberrant binding to this element. We hypothesize that the protein(s) binding to this site contributes to the proper modulation of p21 expression during normal myeloid development.
MATERIALS AND METHODS
Cells and cell separation.
For CD34+ cells, human umbilical cord blood (CB) samples were obtained immediately after delivery in accordance with institutional guidelines, and placed in 50-mL tubes containing ACD-A (Cytosol Labs, Braintree, MA). The CB was diluted with Ca- and Mg-free phosphate-buffered saline (PBS), and low-density mononuclear cells are isolated by Ficoll-Paque (1.077 g/mL) density gradient centrifugation for 30 minutes at 400g (Pharmacia Biochem, Piscataway, NJ). CB mononuclear cells were washed twice in PBS and resuspended in PBS + 0.6% ACD-A for magnetic labeling and separation. CD34+progenitor cells were isolated by immunomagnetic selection techniques, as previously described.36 Briefly, cells were incubated with blocking reagent (human IgG) and QBEND/10 anti-CD34 antibody for 15 minutes at 4°C and then washed in PBS/ACD-A followed by incubation with a secondary antibody-magnetic microbead conjugate for an additional 15 minutes at 4°C. The percentage of CD34+ cells was determined by flow cytometric analysis and generally exceeded 95%.
CD34+ cells were cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with 15% heat-inactivated fetal bovine serum (FBS; Life Technologies, Grand Island, NY) containing 100 ng/mL kit ligand (R&D Systems, Minneapolis, MN), 100 ng/mL interleukin-3 (IL-3; R & D Systems), and 30 ng/mL granulocyte colony-stimulating factor (G-CSF; Neupogen; Amgen, Inc, Thousand Oaks, CA). Parallel experiments in which the cells were grown in serum-free CD34+ supportive media (StemPro TM; Life Technologies) gave essentially the same results. Neutrophils were harvested after spinning plasma and removing residual erythrocytes with hypotonic washes, followed by Ficoll-Paque (1.077 g/mL) density gradient centrifugation for 30 minutes at 400g and collection of the pellet in PBS. Purity (generally >95%) was verified by Wright-Giemsa staining. HL-60 cells were cultured in RPMI media (Mediatech, Washington DC) supplemented with 10% heat-inactivated FBS (Life Technologies Laboratories) and 100 U/mL of penicillin and streptomycin. Cells were split at 1:10 every 4 to 5 days to maintain a concentration of less than 8 × 106/mL. All cells were maintained in log-phase at 37°C in a humidified atmosphere with 5% CO2, and fresh vials from a common freeze were thawed every 4 to 6 weeks to prevent senescence.
Reagents and probes.
Reagents included 12-O-tetradecanoylphorbol 13-acetate (TPA; stored at 800 μmol/L in PBS and diluted for use), butyrate, trans-retinoic acid (stored as a 1 mmol/L stock in alcohol), and dimethylsulfoxide (all from Sigma Chemicals, St Louis, MO) and other reagents from commercial vendors. Full-length p21 probe was obtained by digestion and gel purification from a p21 (WAF1/CIP1) plasmid (kindly provided by Dr Wafik El-Diery, University of Pennsylvania, Philadelphia, PA). All probes were radiolabeled to 5 to 10 × 108 cpm/μg by random priming according to manufacturer's instructions (Life Technologies, Gaithersberg, MD).
Northern blot analysis.
Total RNA was extracted from cell lines by solubilization in 4 mol/L guanidium isothiocyanate followed by centrifugation through a cesium chloride cushion. Fifteen to 30 μg of total RNA was separated on agarose-formaldelhyde gels blotted onto Zetabind (AMF-Cuno, Meridan, CT), covalently bound by brief UV irradiation, and hybridized by established procedures to radiolabeled probes for 12 to 24 hours. Blots were exposed to Kodak XAR-5 film (Eastman Kodak, Rochester, NY) with intensifying screens at −80°C. Exposure times varied from 8 days for p53 probes, 1 to 3 days for p21 probes, and 1 to 12 hours for 18S probes.
In situ hybridization.
Digoxigenin-labeled DNA probes were prepared by random priming using the Genius System and hybridized to cytocentrifuged preparations as described (Boehringer Mannheim, Indianapolis, IN). Briefly, cells were centrifuged onto gelatin-subbed slides, fixed in 4% paraformaldehyde, ethanol-dehydrated, air-dried, and stored at −20°C. Cytospin preparations were prehybridized in 10 mmol/L Tris-HCl, 50% formamide, 0.6 mol/L NaCl, 1 mmol/L EDTA, 1× Denhardt's, 0.5 mg/mL carrier RNA, and 10% dextran sulfate for 1 hour at 37°C. The digoxigenin-labeled p21 probe, diluted in prehybridization buffer, was applied to the cells and allowed to hybridize overnight at 37°C. After posthybridization washes, detection of digoxigenin-labeled probe was accomplished by incubating the cells with alkaline phosphatase-conjugated anti-digoxigenin antibody (1:500) for 2 hours at room temperature Alkaline phosphatase activity was then visualized by incubating the cells in the absence of light with NBT and BICP (Boehringer Mannheim). The color development was monitored and the enzymatic reaction was stopped by immersing the slides in 10 mmol/L Tris, 1 mmol/L EDTA.
Immunostaining.
Cytospins of cells were prepared, dried, and stained indirectly for p21 expression. In brief, cells were fixed with methanol/acetone and incubated in primary antibody (either mouse monoclonal anti-p21 antibody CP36 or isotype control mouse IgG) in 0.2% NP-40 at a 1:50 dilution for 2 hours, followed by four PBS washes and incubation for 30 minutes with biotinylated goat antimouse IgG antibody (Vector Laboratories, Burlingame, CA) again followed by three washes. Immunodetection was enabled by a final 30 minutes of incubation with Cy3-conjugated streptavidin (Jackson Immunochemicals, West Grove, PA) and three PBS washes. Cells were counterstained with Hoechst.
Extract preparation and gel-shift assay.
Cells were washed in PBS and whole cell extracts were prepared by resuspending cell pellets in high salt lysis buffer (20 mmol/L HEPES, pH 7.9, 420 mmol/L NaCl, 20 mmol/L NaF, 1 mmol/L Na3Vo4, 1 mmol/L Na4P2O7, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L dithiothreitol [DTT], 1 μg/mL leupeptin, 1 μg/mL aprotinin, 0.5 mmol/L phenylmethyl sulfonyl fluoride [PMSF], and 50% glycerol) followed by three rapid freeze-thaw cycles. For gel-shift assays, equivalent amounts of extract protein were preincubated for 20 minutes in binding buffer (15 mmol/L HEPES, pH 7.9, 65 mmol/L NaCl, 1 mmol/L DTT, 0.15 mmol/L EDTA, 8% glycerol) with poly dI:dC (0.3 μg) followed by 20 minutes of incubation with 0.2 ng end-labeled target sequence, followed by fractionation on a 4% nondenaturing polyacrylamide gel.
Plasmids.
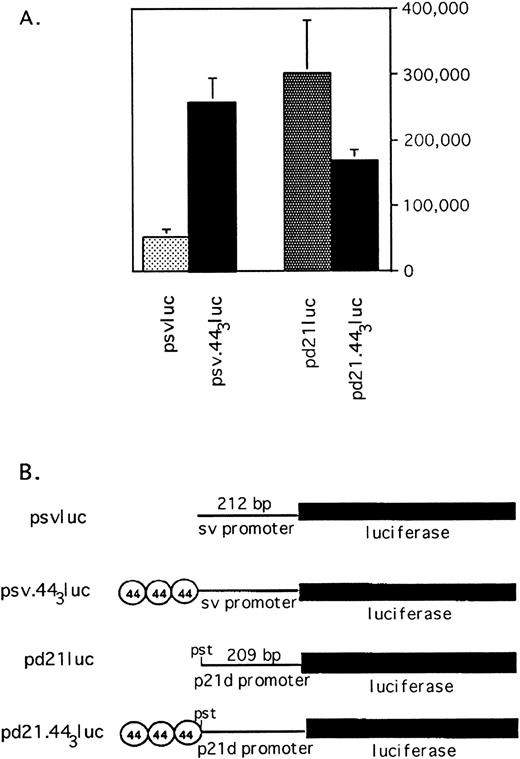
Plasmid psvluc is used to designate pGL2 promoter (Promega, Inc, Seattle WA) that contains an sv40 promoter upstream of the luciferase gene; plasmid p21luc contains 2.4 kb of the p21 promoter cloned into the HindIII site of pGL2basic (Promega, Inc) and was a gift from Dr Xiao-Fan Wang (Duke University Medical Center). Plasmid pd21luc consists of a truncated p21 promoter in which 2.2 kb of p21 promoter upstream sequence was deleted from p21 luc. It was contructed by digesting p21luc with Sac I and Pst I, filling in overhanging ends, and autoligating. Plasmid pd21.443 luc contains a triplication of the 44-bp fragment upstream of that truncated promoter. The 44-bp fragment was synthesized with GATC-overhangs, autoligated, and cloned into theBamHI site of pBluescript, generating pBlue-443. ASac I-Pst I fragment from pBlue-443 was cloned into pd21 to generate pd21.443luc. Plasmid psv.443luc similarly contains a triplication of 44-bp fragments cloned into a Sac I-Xho I site upstream of the sv40 promoter.
Transient transfection.
Log-phase HL-60 were washed twice in Opti-mem medium (Life Technologies) and 107 cells were suspended in 200 μL Opti-mem per transfection. A total of 15 μg of uncut plasmid (reporter plus transfection control) in 50 μL Opti-mem medium was added to the cells and the mixture was preincubated in a chilled electroporation cuvette for 10 minutes. Electroporation was performed in a Bio-Rad electroporation apparatus (Bio-Rad, Hercules, CA) at 270 V, 960 μF, generating a time constant around 70. The mixture was maintained on ice for 10 minutes followed by dilution in RPMI medium containing 10% FBS. Cells were harvested 6 hours later and lysed by freeze-thaw cycles, and luciferase activity and β-galactosidase activity were determined by standard techniques on aliquots of 20% of the extract.
RESULTS
Stage-specific expression of p21 in differentiating myeloid cells.
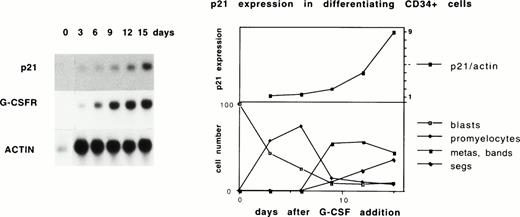
We have previously demonstrated that p21 expression is directly upregulated in HL-60 cells by multiple chemical inducers of differentiation. This upregulation occurs at the mRNA and protein levels and does not require new protein synthesis because it is not blocked by cycloheximide.16 Other p53-negative hematopoietic and hepatoma cell lines displayed similar upregulation of p21 within hours of the addition of differentiation inducers.16 To assess whether similar induction of p21 occurred during progressive differentiation of normal hematopoietic cells, p21 message was determined in differentiating CD34+cells. CD34+ cells were harvested from umbilical cord blood, expanded for 3 days with kit-ligand and IL-3, and then driven to differentiate along the myeloid lineage by addition of G-CSF. Such an approach has been shown37 to lead to roughly synchronous differentiation. Figure 1 demonstrates progressive upregulation of p21 as the cells differentiate. Nonadherent cells were collected at 3-day intervals after the addition of G-CSF and analyzed for morphology and for p21 message on Northern blot. Representative results of triplicate experiments are shown. Concurrent upregulation of G-CSF receptor confirms granulocytic differentiation.38 The sharpest increase in p21 levels occurs coincident with the appearance of metamyelocytic and granulocytic cells on cytospin. These data establish a progressive increase in p21 message as cells mature.
p21 upregulation during myeloid differentiation of CD34+ cells. CD34+ cells were expanded in 100 ng/mL IL-3 and 100 ng/mL KL in 10% IMDM for 3 days and then G-CSF was added at 10 ng/mL. Similar results are seen if G-CSF is added at day 0. At day 0 and at 3-day intervals after the addition of G-CSF, nonadherent cells were harvested for morphologic analysis and RNA preparation. (Right) (bottom) Morphologic analysis of 86-107 cells on cytospins shown as percentages; myelocytes (not shown) peaked at 6%. (Top) Normalized p21 message as determined on a phosphoimager is plotted. (Left) RNA was blotted and sequentially probed with radiolabeled p21, G-CSF receptor, and actin cDNAs.
p21 upregulation during myeloid differentiation of CD34+ cells. CD34+ cells were expanded in 100 ng/mL IL-3 and 100 ng/mL KL in 10% IMDM for 3 days and then G-CSF was added at 10 ng/mL. Similar results are seen if G-CSF is added at day 0. At day 0 and at 3-day intervals after the addition of G-CSF, nonadherent cells were harvested for morphologic analysis and RNA preparation. (Right) (bottom) Morphologic analysis of 86-107 cells on cytospins shown as percentages; myelocytes (not shown) peaked at 6%. (Top) Normalized p21 message as determined on a phosphoimager is plotted. (Left) RNA was blotted and sequentially probed with radiolabeled p21, G-CSF receptor, and actin cDNAs.
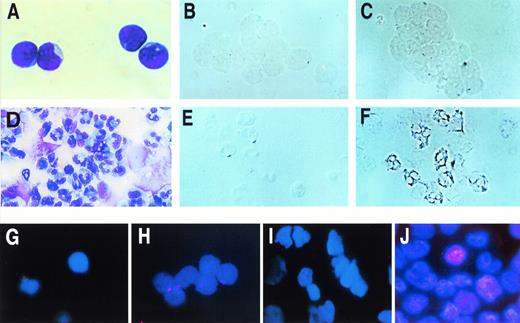
The absence of p21 message in CD34+ blast cells and presence of p21 message in neutrophils was also demonstrated by in situ hybridization (Fig 2A through F). To characterize the variation in p21 expression within this mixed population of differentiating cells, cytospins of undifferentiated or differentiating cells were stained for p21 expression. It is evident that little p21 protein is expressed by the CD34+ cells before culture in cytokines (Fig 2G and H) and that p21 is expressed in most cells after 11 days of differentiation, with high levels of expression in a subset of cells (Fig 2I and J).
p21 message and protein in CD34+ blast cells and in differentiated cells. (Top) In situ analysis of p21 message in CD34+ blast cells (above) and in neutrophils (below). (A) and (D), Wright Giemsa; (B) and (E), digoxigenin-labeled control; (C) through (F), digoxigenin-labeled p21 probe. (Bottom) Immunohistochemical demonstration of p21 protein in differentiating cells. Day 0 CD34+ blasts stained with isotype control (G) or anti-p21 antibodies. Some nonspecific staining of antibody-coated magnetic beads is present. Differentiated progeny arising after 11 days of culture stained with isotype control (I) or anti-p21 antibody (J) stain strongly for p21.
p21 message and protein in CD34+ blast cells and in differentiated cells. (Top) In situ analysis of p21 message in CD34+ blast cells (above) and in neutrophils (below). (A) and (D), Wright Giemsa; (B) and (E), digoxigenin-labeled control; (C) through (F), digoxigenin-labeled p21 probe. (Bottom) Immunohistochemical demonstration of p21 protein in differentiating cells. Day 0 CD34+ blasts stained with isotype control (G) or anti-p21 antibodies. Some nonspecific staining of antibody-coated magnetic beads is present. Differentiated progeny arising after 11 days of culture stained with isotype control (I) or anti-p21 antibody (J) stain strongly for p21.
Loss of a p21 promoter-binding activity during differentiation.
We reasoned that stage-specific regulation of p21 expression was likely to be a central feature of the physiologic role of p21 and therefore might be shared across species. Should that be the case, homologous sequences within the p21 promoter that bind similar regulatory factors might be conserved. Comparison of the sequences of the human, mouse, and rat p21 promoter39 disclosed a striking 44-bp region of homology (bp −1367 to −1324 of the human promoter). This region also contains two sequence elements present in the p27 promoter:
There are no other high-level matches to the 44-bp sequence on BLAST analysis of the Genbank database. There are several sequence features in this fragment that could be of significance. The fragment contains a motif, TTN5AA, which conceivably could be recognized by Stat proteins. There is also a hexameric consensus motif (AGGTCA) for orphan nuclear receptors40 and a recognition site for the pu.1 transcription factor. The fragment contains a direct repeat of a sequence (CTGGGCAT/G) that is present six times in the p21 promoter. It is noteworthy that a putative p53 binding sequence (italicized) is contained within this fragment.23,39,41 To determine whether proteins other than p53 bound to this region, we assayed HL-60 extracts for binding activity to this 44-bp fragment. It should be noted that HL-60 cells are p53-negative due to homozygous deletions42 and do not express p53. Any activity in protein extracts of HL-60 binding to this fragment would therefore be distinct from p53. As shown in Fig 3, HL-60 cells express a protein that binds specifically to this p21 promoter fragment. This activity will hereafter be referred to as 21PBA (p21 promoter-binding activity). Binding is eliminated by specific but not by nonspecific competitor DNA. Both target strands are required for binding, which is eliminated by protease treatment (data not shown).
HL-60 cells express a DNA-binding activity that specifically binds a 44-bp fragment in the p21 promoter. Whole cell extracts of log-phase HL-60 cells were incubated in the presence of a radiolabeled 44-bp fragment from the p21 promoter and subjected to electrophoretic mobility shift analysis. Binding was performed in the presence of 30× or 100× excess of specific (44) or non-specific (YY1) unlabeled competitor DNA as indicated.
HL-60 cells express a DNA-binding activity that specifically binds a 44-bp fragment in the p21 promoter. Whole cell extracts of log-phase HL-60 cells were incubated in the presence of a radiolabeled 44-bp fragment from the p21 promoter and subjected to electrophoretic mobility shift analysis. Binding was performed in the presence of 30× or 100× excess of specific (44) or non-specific (YY1) unlabeled competitor DNA as indicated.
Binding during differentiation of HL-60 and CD34+ cells.
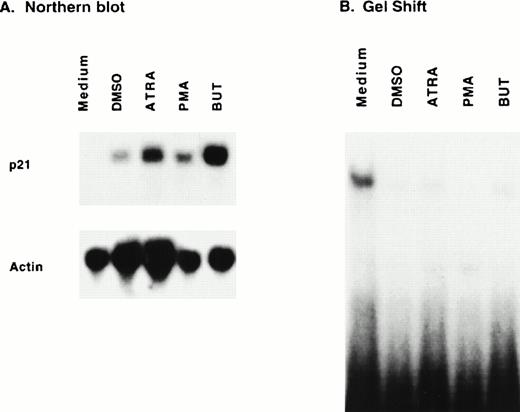
To see whether proteins binding to this sequence were expressed in a stage-specific manner during differentiation, we assayed extracts of exponentially growing HL-60 cells and of HL-60 cells differentiated with various agents for 21PBA. Exponentially growing HL-60 cells were incubated with chemical inducers of differentiation, and RNA was extracted 20 hours later. Northern blot analysis (Fig 4A) confirmed upregulation of p21 message by all inducers used. The ability of proteins present in undifferentiated and differentiated cell extracts to bind to the 44-bp fragment was determined using a gel shift assay. Figure 4B demonstrates that proliferating HL-60 cells contain a DNA-binding activity, and that binding activity is decreased in extracts of differentiating HL-60 cells.
Protein-binding of a p21 promoter fragment is inversely associated with p21 message expression. Log-phase HL-60 cells were exposed to medium alone or medium containing DMSO (1.25%), ATRA (1 μmol/L), PMA (80 nmol/L), or butyrate (1 mmol/L) and harvested 20 hours later for RNA and protein extraction. Northern blot (A) confirms p21 induction. Gel-shift assay (B) of 20 μg whole cell extract incubated with 44-bp promoter fragment discloses a band upon addition of uninduced extract, which is decreased in lanes using differentiating cell extract.
Protein-binding of a p21 promoter fragment is inversely associated with p21 message expression. Log-phase HL-60 cells were exposed to medium alone or medium containing DMSO (1.25%), ATRA (1 μmol/L), PMA (80 nmol/L), or butyrate (1 mmol/L) and harvested 20 hours later for RNA and protein extraction. Northern blot (A) confirms p21 induction. Gel-shift assay (B) of 20 μg whole cell extract incubated with 44-bp promoter fragment discloses a band upon addition of uninduced extract, which is decreased in lanes using differentiating cell extract.
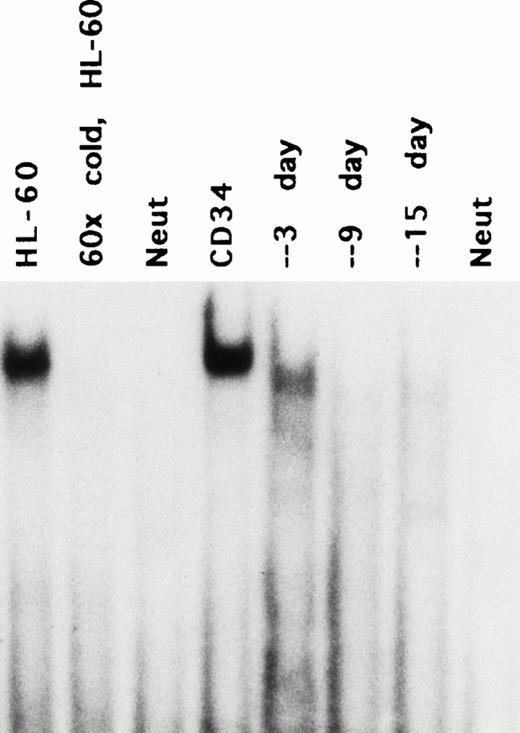
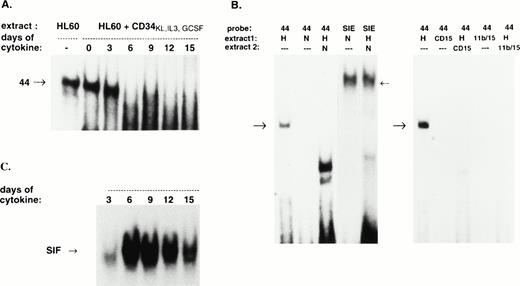
To determine whether 21PBA existed in normal hematopoietic precursor cells induced to undergo myeloid differentiation, extracts were prepared from umbilical cord CD34+ cells upon harvest and at different times after culture in differentiating medium consisting of kit ligand, IL-3, and G-CSF. As shown in Fig 5, CD34+ cells also contained a DNA-binding protein that recognized the same fragment and yielded the same gel shift as in HL-60 cells. As in HL-60 cells, the 21PBA was extinguished as CD34+ cells differentiated and p21 levels increased; binding was undetectable 6 days after the cells had been induced to differentiate. In contrast, DNA binding proteins targeting other elements were active in the differentiating CD34+ cell extracts, as demonstrated by their ability to bind a serum inducible element target (SIE; GATCCATTTCCCGTAAATCGATC43) on gel shift assay (see Fig 7C).
Stage-specific presence of p21-promoter binding activity in differentiating CD34+ cells. Umbilical cord CD34+ cells prepared with magnetic beads were expanded in differentiating medium (KL, IL-3, and G-CSF) and extracts prepared for gel-shift in parallel with p21 mRNA determination as shown in Fig 1. The faint and slightly faster band at day 3 is reproduceable. Lane 2 indicates competition by cold fragment of the band induced by HL-60 extract.
Stage-specific presence of p21-promoter binding activity in differentiating CD34+ cells. Umbilical cord CD34+ cells prepared with magnetic beads were expanded in differentiating medium (KL, IL-3, and G-CSF) and extracts prepared for gel-shift in parallel with p21 mRNA determination as shown in Fig 1. The faint and slightly faster band at day 3 is reproduceable. Lane 2 indicates competition by cold fragment of the band induced by HL-60 extract.
Elimination DNA-binding activity of extracts by coincubation with differentiated cell extracts. (A) Twelve micrograms of HL-60 extract was incubated in the absence (−) or presence of 12 μg of extract from CD34+ blast cells (0) or CD34+ cells incubated for 3 to 15 days in the presence of kit ligand (100 ng/mL), IL-3 (100 ng/mL), and G-CSF (20 ng/mL) as indicated. After 20 minutes of coincubation, radiolabeled target DNA was added and gel-shift was performed as described. (B) (Left half) Thirty micrograms of extract from HL-60 cells (H) and cord neutrophils (N) were incubated separately or together (H+N) before the addition of labeled 44-bp fragment or labeled SIE probes. The typical gel-shift band seen upon incubation with each fragment is indicated with an arrow (forward arrow, 44-bp band shift; backward arrow, SIF bands). The lower, nonspecific band seen in the H+N lane was not extinguished by unlabeled competitor 44-bp fragment (not shown). (Right half) Twenty-six micrograms of HL-60 extract (H) or 9 μg of extract from sorted CD11b−CD15+ cells (CD15) or from CD11B+CD15+ cells (11b/15) were incubated separately with labeled 44-bp fragment as shown. In addition, these amounts of extracts were combined in mixing experiments (H+CD15), (H+11b/15) as indicated. Specific bands are indicated as before. (C) Incubation of differentiating cell extracts with SIE. Extracts from CD34+ cells incubated for 3 to 15 days in the presence of kit ligand, IL-3, and G-CSF were incubated with radiolabeled SIE target. Characteristic bands (SIF43) are evident.
Elimination DNA-binding activity of extracts by coincubation with differentiated cell extracts. (A) Twelve micrograms of HL-60 extract was incubated in the absence (−) or presence of 12 μg of extract from CD34+ blast cells (0) or CD34+ cells incubated for 3 to 15 days in the presence of kit ligand (100 ng/mL), IL-3 (100 ng/mL), and G-CSF (20 ng/mL) as indicated. After 20 minutes of coincubation, radiolabeled target DNA was added and gel-shift was performed as described. (B) (Left half) Thirty micrograms of extract from HL-60 cells (H) and cord neutrophils (N) were incubated separately or together (H+N) before the addition of labeled 44-bp fragment or labeled SIE probes. The typical gel-shift band seen upon incubation with each fragment is indicated with an arrow (forward arrow, 44-bp band shift; backward arrow, SIF bands). The lower, nonspecific band seen in the H+N lane was not extinguished by unlabeled competitor 44-bp fragment (not shown). (Right half) Twenty-six micrograms of HL-60 extract (H) or 9 μg of extract from sorted CD11b−CD15+ cells (CD15) or from CD11B+CD15+ cells (11b/15) were incubated separately with labeled 44-bp fragment as shown. In addition, these amounts of extracts were combined in mixing experiments (H+CD15), (H+11b/15) as indicated. Specific bands are indicated as before. (C) Incubation of differentiating cell extracts with SIE. Extracts from CD34+ cells incubated for 3 to 15 days in the presence of kit ligand, IL-3, and G-CSF were incubated with radiolabeled SIE target. Characteristic bands (SIF43) are evident.
Aberrant binding in leukemia cells.
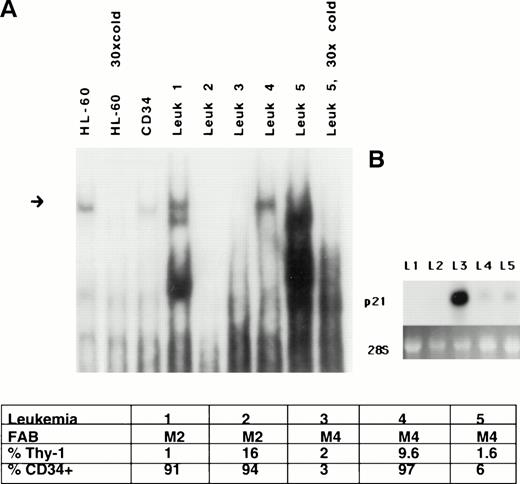
Aberrant expression of p21 protein has been reported in leukemia, and dysregulated expression of p21 has been posited to be a negative prognostic sign.44 Because of this observation and because normal differentiation is dysregulated in leukemia, we analyzed the expression of 21PBA in leukemic blasts freshly harvested from untreated patients. Figure 6 demonstrates that leukemic extracts vary in their ability to bind to the p21 promoter fragment. Five AML leukemic samples varied both in the presence of binding activity and in the patterns of bands arising on gel-shift assay. There was no clear correlation between binding activity and CD34+ percentage or French-American-British (FAB) class. Expression of p21 message was also determined. It is apparent that the expression of p21 in leukemic cells is complex. Patient samples no. 3 and 5, classified as M4 leukemias, express similar low levels of CD34+ and Thy-1, but sample no. 3 lacks 21PBA activity and expresses high levels of p21 transcripts, whereas sample no. 5 expresses high levels of 21PBA and low expression of p21 message. However, it is clear that additional factors regulate p21 in leukemic cells. This is evident in sample no. 2, in which both 21PBA and p21 message are undetectable.
Altered p21-promoter binding in a subset of leukemia cell extracts. (A) Protein extracts were prepared from five random frozen samples of leukemic blasts from peripheral blood of patients presenting acutely with AML, and binding to the p21-promoter was determined on gel shift. Differences in the presence and in the mobility of binding complexes are evident. The slowly migrating band in extract from patient no. 5 is specific and competed by cold fragment. A short exposure is shown. HL-60 and CD34 extracts are loaded for comparison. Equal amounts of protein are used in each binding reaction. FAB and FACS analysis results of the samples are also shown. (B) Northern blot demonstrates variable expression of p21 message in the leukemic samples.
Altered p21-promoter binding in a subset of leukemia cell extracts. (A) Protein extracts were prepared from five random frozen samples of leukemic blasts from peripheral blood of patients presenting acutely with AML, and binding to the p21-promoter was determined on gel shift. Differences in the presence and in the mobility of binding complexes are evident. The slowly migrating band in extract from patient no. 5 is specific and competed by cold fragment. A short exposure is shown. HL-60 and CD34 extracts are loaded for comparison. Equal amounts of protein are used in each binding reaction. FAB and FACS analysis results of the samples are also shown. (B) Northern blot demonstrates variable expression of p21 message in the leukemic samples.
Suppression of promoter-binding activity by extracts from neutrophils or from differentiating CD34+ cells.
The loss of 21PBA in differentiated cells could arise from lack of production or from lack of activation of necessary binding factors in those cells or from sequestration of binding factors into other complexes, making them unavailable to form a binding complex on the p21 promoter. To clarify this, extract mixing experiments were performed in which extracts from neutrophils or differentiated CD34+cells were mixed with uninduced HL60 cell extracts before assaying the HL60 extracts for gel-shift activity. Figure 7A demonstrates that CD34+ extracts from cells exposed to G-CSF for 6 or more days suppressed the binding activity present in extracts of exponentially growing HL-60 cells. To demonstrate that the absence of the competed band in Fig 7A does not result from nonspecific inhibitors in the competing extracts from the differentiating cells, those extracts were incubated separately with a serum-inducible element target sequence (SIE). Figure 7C demonstrates the ability of the competing extracts to generate SIF bands43 upon incubation with an SIE target, indicating their patency. Extracts from neutrophils suppressed the ability of HL-60 extracts to bind the 44-bp p21 promoter fragment (Fig 7B, left panel). Neutrophil extract also suppressed 44-bp binding by extract from CD34+ blast cells (data not shown). Suppressive activity was heat-labile and exhibited by both whole cell and nuclear extracts of neutrophils (data not shown). Experiments represented in the right panel of Fig 7B tested whether extracts from distinct subpopulations of differentiating CD34+ cells differed in their ability to suppress the 44-bp fragment binding by uninduced HL-60 extracts. CD34+ blasts were induced to differentiate for 9 days, and two subpopulations of cells were separately collected by fluorescence-activated cell sorting (FACS). One set of cells expressed CD15 antigen but not the CD11b antigen; the other, more mature population expressed both CD11b and CD15 antigens. Gel-shift analysis demonstrated that neither of these cell types exhibited binding activity for the 44-bp fragment, and extracts from either population could inhibit the binding activity present in HL-60 extracts.
The 44-bp p21 promoter fragment is transcriptionally active.
The existence of proteins in CD34+ blast cells and HL-60 cells that specifically recognized and bound to the 44-bp fragment within the p21 promoter raised the prospect that this fragment was transcriptionally active. Moreover, the inverse association of DNA-binding and p21 mRNA expression suggested that a transcriptional repressor bound to the p21 promoter at this site. Figure 8 shows the results of transfection experiments using several constructs to evaluate the transcriptional activity of the 44-bp element. This activity was assessed upstream of heterologous promoters. This avoids the confounding effects inherent in testing within the full-length p21 promoter. Stress-responsive elements within the full-length p21 promoter activate transcription in response to transfection, masking the effects of upstream mutations (Timchenko et al45 and R.S., unpublished observations). These transfections were performed using the p53-negative HL-60 cell line to avoid confounding effects of p53 on the transcriptional activity of the 44-bp fragment.
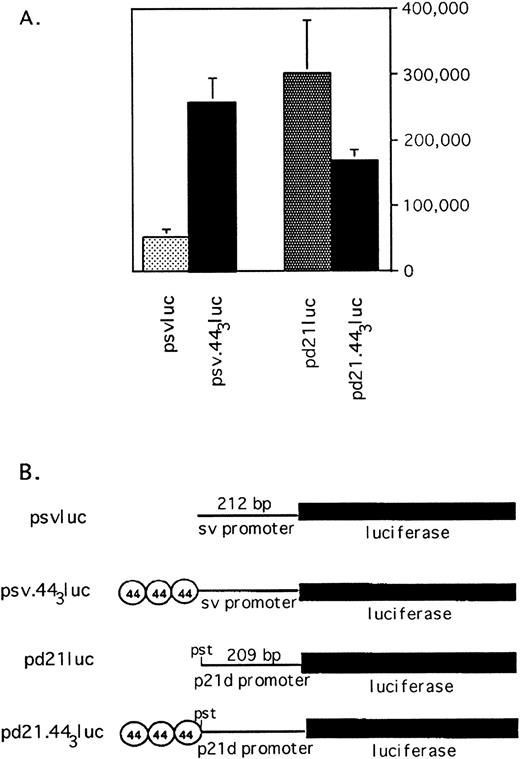
p53-independent transcription mediated by the 44-bp element. (A) The 44-bp element enhances or suppresses transcription depending on the promoter. Log phase HL-60 cells were transfected with luciferase reporter plasmid driven by the sv40 promoter (psvluc) or a promoter consisting of 209 bp of the p21 promoter proximal to the transcription startsite (pdluc). Luciferase activity is compared with that of reporter constructs with 3 copies of the 44-bp sequence upstream of the promoter, as indicated. All values are normalized to β-galactosidase expression from cotransfected CMV-βgal reporter to correct for transfection efficiency. The average and standard error of three experiments is shown. (B) Schematic of reporter plasmids used in transfections.
p53-independent transcription mediated by the 44-bp element. (A) The 44-bp element enhances or suppresses transcription depending on the promoter. Log phase HL-60 cells were transfected with luciferase reporter plasmid driven by the sv40 promoter (psvluc) or a promoter consisting of 209 bp of the p21 promoter proximal to the transcription startsite (pdluc). Luciferase activity is compared with that of reporter constructs with 3 copies of the 44-bp sequence upstream of the promoter, as indicated. All values are normalized to β-galactosidase expression from cotransfected CMV-βgal reporter to correct for transfection efficiency. The average and standard error of three experiments is shown. (B) Schematic of reporter plasmids used in transfections.
Figure 8A demonstrates the effect of the 44-bp repeat on heterologous promoters sv and pd21dluc. These plasmids or the corresponding plasmids bearing 44-bp repeats upstream of the promoters were transfected into HL-60 cells along with CMV-βgal as a transfection efficiency control. Transcription from the sv-promoter is enhanced fivefold by upstream placement of a triplication of the 44-bp sequence. In contrast, insertion of 44-bp repeats upstream of the truncated p21-promoter decreases transcription roughly twofold, indicating that the transcriptional effects of this element are promoter-specific, as has been shown for other enhancer/repressor elements.46-48
Novelty of 21PBA binding.
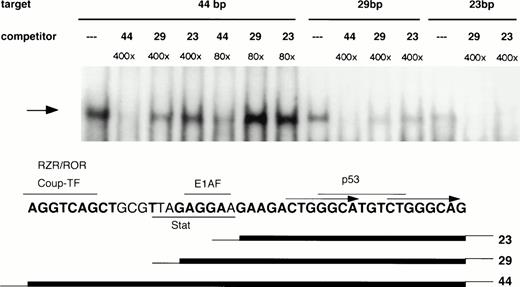
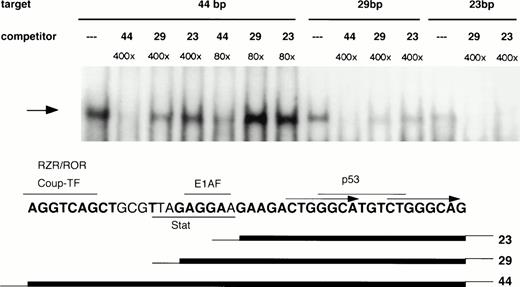
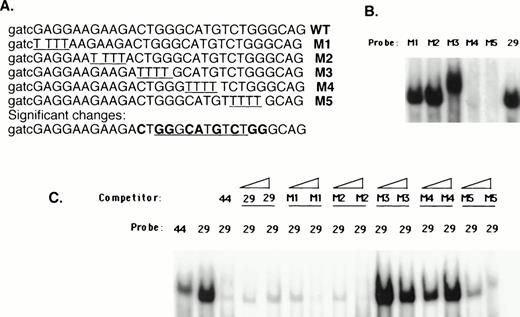
Mapping of nucleotides involved in 21PBA binding was undertaken to clarify whether this activity might represent a known transcription factor. The upstream half of the 44-bp element contains consensus binding sequences for orphan nuclear receptors RZR/ROR or Coup-TF,40,49,50 the E1AF51 Ets-family transcription factor, and a potential Stat binding site. These factors have been shown to affect p21 expression, although binding within this sequence has not been established for any of them. The downstream 22-bp portion of the 44-bp element lacks major transcription factor binding sequences, with the exception of a p53 consensus binding sequence (GGGCATGTCT; Fig 9). To determine whether 21PBA represents one of these known factors or might conceivably represent a novel factor, we first determined whether 21PBA bound to the upstream or downstream half of the fragment. As is shown in Fig 9, both a 29-bp and 23-bp truncation of the 44-bp element gave rise to the same band on gel-shift assays as the full-length fragment, suggesting that sequences within the downstream half of that fragment comprised a recognition site for 21PBA. However, neither a 400-fold excess of 29 bp (−1351 to −1323) or 23 bp (−1345 to −1323) effectively competed the full-length fragment, indicating that these shorter fragments did not result in the highest affinity binding. This suggests that binding of 21PBA is stabilized by sequences present in the 5′-half of the full-length 44-bp fragment. To more precisely to map nucleotides required for 21PBA binding, scanning mutations were made in the 28-bp fragment (Fig 10A) in which successive 4-bp sequences were replaced by thymidines. The third (M3) and fourth (M4) substitutions negated ability of mutant sequence to compete binding with wild-type; the fifth (M5) substitution retained partial competitive ability (Fig 10B). Consistent with these findings, binding of HL-60 extracts to the M4 and M5 mutants was markedly decreased (Fig 10C); the M3 target resulted in altered mobility, which most likely reflects impaired DNA-protein complex formation. The scanning mutation results are also supported by our observation that bisecting the 44-bp fragment at bp −1336 (ACT↑GG↓) eliminated binding (data not shown).
Boundaries of the 21PBA target sequence. 21PBA binds the 3′-half of the 44-bp promoter fragment. HL-60 cell extract was incubated with either the 44-bp fragment or 23- or 29-bp fragments bearing identical sequence to the 3′ portion of the longer fragment. All targets generate the same gel-shift band, which can be specifically competed, as shown. The 44-bp fragment is significantly more effective than the shorter fragments as a cold competitor against labeled 44- or 28-bp fragments. The bottom portion of the figure depicts the positions of recognition sites for several transcription factors relative to fragment boundaries.
Boundaries of the 21PBA target sequence. 21PBA binds the 3′-half of the 44-bp promoter fragment. HL-60 cell extract was incubated with either the 44-bp fragment or 23- or 29-bp fragments bearing identical sequence to the 3′ portion of the longer fragment. All targets generate the same gel-shift band, which can be specifically competed, as shown. The 44-bp fragment is significantly more effective than the shorter fragments as a cold competitor against labeled 44- or 28-bp fragments. The bottom portion of the figure depicts the positions of recognition sites for several transcription factors relative to fragment boundaries.
Scanning mutation analysis of 21PBA recognition sequence. (A) Juxtaposition of mutated sequences M1-M5 against the wild-type sequence. Successive 4-bp sequence elements are substituted with thymidine. Base changes affecting 21PBA binding are summarized at the bottom in bolded letters; sequences comprising a p53 consensus site are underlined. (B) Binding by mutant sequences is shown. Equal amounts of radiolabeled mutant (M1-M5) or wild-type (29) probe were incubated with HL-60 extract in gel-shift assay. Loss of binding to M4 and M5 and novel migration using the M3 target is evident. (C) The ability of mutant sequences to compete binding to the 29-bp target is shown. A 44-bp target labeled at low activity is shown in lane 1. HL-60 extract is incubated with radiolabeled 29-bp target after preincubation for 30 minutes with 30-fold excess of unlabeled 44-bp competitor and either 30- or 100-fold excess of unlabeled 29-bp or mutant competitor as shown.
Scanning mutation analysis of 21PBA recognition sequence. (A) Juxtaposition of mutated sequences M1-M5 against the wild-type sequence. Successive 4-bp sequence elements are substituted with thymidine. Base changes affecting 21PBA binding are summarized at the bottom in bolded letters; sequences comprising a p53 consensus site are underlined. (B) Binding by mutant sequences is shown. Equal amounts of radiolabeled mutant (M1-M5) or wild-type (29) probe were incubated with HL-60 extract in gel-shift assay. Loss of binding to M4 and M5 and novel migration using the M3 target is evident. (C) The ability of mutant sequences to compete binding to the 29-bp target is shown. A 44-bp target labeled at low activity is shown in lane 1. HL-60 extract is incubated with radiolabeled 29-bp target after preincubation for 30 minutes with 30-fold excess of unlabeled 44-bp competitor and either 30- or 100-fold excess of unlabeled 29-bp or mutant competitor as shown.
The 21PBA recognition sequence overlaps a p53 binding sequence.
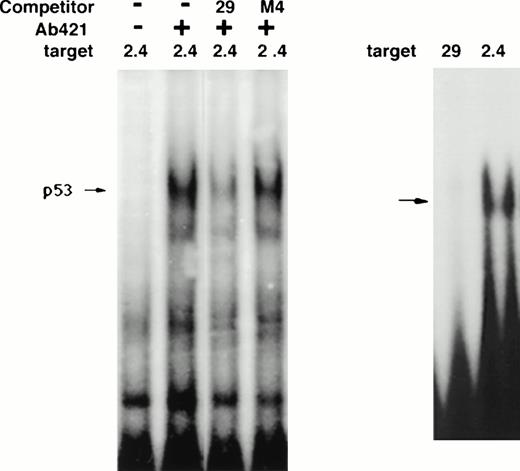
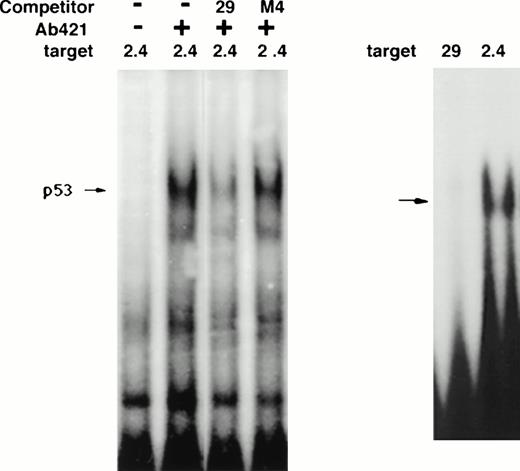
As Fig 10 demonstrates, 21PBA recognition site consists of a downstream 16-bp sequence ACTGGGCATGTCTGGG (bp −1340 to −1325). Bases changed in the mutation sets 3, 4, and 5 are in bold; these mutations disrupt the p53 consensus and also inhibit 21PBA binding. Analysis of this sequence using the MatInspector52 and Transfac53 analysis programs identify no known transcriptional factor binding sites within this sequence; MacVector analysis highlights the p53 binding sequence PuPuPuC(A/T)(T/A)GPyPyPy (underlined within the 16-bp shown above; optimal p53 binding requires a repeat of this sequence54-56). The p53 site overlap includes the bases (CATG) essential for 21PBA binding that are altered in mutation 4. Despite the overlap in recognition sequence, 21PBA is clearly not p53, because it is present in p53-negative HL-60 cells as well as in other cells lacking wt p53, including K562, HT-29, and C33A (data not shown). Although this p53 binding sequence has been reported to be bound weakly57 by p53 using in vivo footprinting and in a sensitive immunoprecipitation assay,23 it has not been shown to bind p53 in gel-shift assays. Indeed, we have not detected binding of a recombinant p53 core binding protein58 to the 29-bp sequence under conditions in which it binds to the upstream p53-binding site of the p21 promoter (Fig11). Similar results were seen with cell extracts containing activated p53 (data not shown). The wild-type 29-bp sequence is able to compete binding of p53 to a p53 site containing two PuPuPuC(A/T)(T/A)GPyPyPy repeats (the p53 site 2.4 kb upstream of the p21 transcriptional startsite, labeled 2.4 in Fig 11). In contrast, the M4 mutant sequence is unable to compete p53 binding, underscoring the importance of the mutated bases for p53 binding as well as for 21PBA binding.
p53 binding ability of 29-bp sequence. (A) Ability of 29-bp sequence to compete p53 binding in extracts. Extracts of irradiated T10 cells are tested on gel-shift assay. The upstream p53 site in the p21 promoter (2.4) is targeted. The first two lanes demonstrate the appearance of the expected p53 gel-shift band in the presence of anti-p53 antibody pAb421. The p53 band is partially competed by 100-fold excess of 29-bp sequence but not mutated sequence (M4). (B) Lack of direct binding of p53 to the 29-bp sequence. A gel-shift assay is shown demonstrating binding of recombinant p53 core protein to the upstream p53 site (2.4) but not to the 29-bp sequence.
p53 binding ability of 29-bp sequence. (A) Ability of 29-bp sequence to compete p53 binding in extracts. Extracts of irradiated T10 cells are tested on gel-shift assay. The upstream p53 site in the p21 promoter (2.4) is targeted. The first two lanes demonstrate the appearance of the expected p53 gel-shift band in the presence of anti-p53 antibody pAb421. The p53 band is partially competed by 100-fold excess of 29-bp sequence but not mutated sequence (M4). (B) Lack of direct binding of p53 to the 29-bp sequence. A gel-shift assay is shown demonstrating binding of recombinant p53 core protein to the upstream p53 site (2.4) but not to the 29-bp sequence.
DISCUSSION
This report demonstrates progressive upregulation of p21 message and protein during maturation of hematopoietic progenitor cells. Previous studies by ourselves and others had established an association between differentiation and p21 expression in cell line models. This report, as well as immunohistochemical evidence that p21 expression is high in differentiated tissues,23 24 bolsters the case that p21 contributes to normal differentiation. We present evidence that the transcriptional activation of p21 during myelopoiesis is highly regulated and is associated with protein-binding to a highly conserved sequence in the p21 promoter.
Expression of p21 message increases in CD34+ cells undergoing myelomonocytic differentiation. Because more mature progeny arose sequentially under our culture conditions,37 the sharpest increase in p21 message could be seen to coincide with the emergence of metamyelocytes and neutrophils, cells that do not proliferate.59 This supports a role for p21 in mediating growth arrest in differentiated cells.16,17,60 However, p21 is clearly present at earlier time points characterized by proliferating cells. This is consistent with the notion that p21 may serve other functions, such as preventing apoptosis,25-28or may play a primary role in differentiation independent of its effects on the cell cycle.61 It has been reported that p21 action is stoichiometric and that the antiproliferative action of p21 requires binding of two p21 molecules to the cyclin-cdk complex.62 63 There may therefore be a threshold level at which p21 expression promotes G1 arrest. We have found that differentiating CD34+ cells include subpopulations that are negative, dim, or bright for p21 (Fig 2 and Yaroslavskiy et al, manuscript submitted); full characterization of each population is underway. A distinct range of p21 levels may choreograph the transition from growing precursor cells to a postmitotic, differentiated population.
What regulates the stage-specific alterations in p21 expression in differentiating CD34+ cells? Transcriptional activation of p21 by MyoD22 and p30031 contribute to its upregulation during differentiation of C2 mouse myoblasts. Such findings raise the prospect that similar regulators exist in other systems to activate p21 at appropriate maturational stages. Such a specific regulator of p21 expression during myeloid differentiation has not been elucidated.
Our data in both cell line models and in normal hematopoietic precursors undergoing maturation indicated that a highly conserved 44-bp sequence in the p21 promoter could be a target for a regulator of p21 expression specific to maturation stage. Binding proteins recognizing this sequence were present in blasts and to a lesser degree in promyelocytes and absent thereafter. Disruption of this stage-specific pattern of expression may occur in a setting of abnormal growth and differentiation. Using extracts from primary leukemia cells, gel-shift analysis of proteins binding to the 44-bp fragment results in aberrant gel-shift patterns or in absence of binding in blasts. This suggests that the protein recognizing this sequence may be a target of dysregulation in leukemia.
The fact that binding to this sequence was inversely correlated with p21 message levels in normal CD34+ cells and in differentiating HL-60 cells raised the prospect either that the binding protein acted as a basal transcriptional repressor or that it prevented access by an activating factor. Previous reports indicating induction of p21 by cycloheximide16,64 support the view that p21 transcription is subject to negative modulation. However, transient transfection experiments demonstrated either an activating or a weak repressing function of this fragment when it was placed upstream of heterologous promoters and transfected into HL-60 cells. It has been noted elsewhere that the ability of transcriptionally active factors to function as activators or as repressors is highly dependent on the surrounding DNA region and the environment of surrounding binding sites.46-48 However, tranfection experiments represent an imperfect approximation of the role of these elements in their native context. Indeed, although p21 message is undetectable in growing HL-60 cells, we have noted high levels of luciferase activity of p21-promoter-driven constructs transfected into these cells. In fact, we have noted that the process of transfection upregulates endogenous p21 mRNA on Northern blots in HL-60 cells (R.S., unpublished observations). Similar transfection-mediated upregulation of p21 has been reported elsewhere.45
It is provocative that this differentiation stage-specific binding activity, 21PBA, recognizes a 16-bp DNA sequence that also contains a recognition sequence for p53.23 57 Mutations in the binding site that alter critical nucleotides in the consensus binding sequence for p53 also abolish 21PBA binding. This raises the interesting prospect that occupation of this site by the 21PBA protein precludes binding by p53 to this sequence. Isolation of the 21PBA protein components is underway to enable precise mapping of the overlap of these binding sites and to establish whether a functional relationship exists between 21PBA and p53 binding.
An interesting aspect of this study has been the observation that differentiated cell extract can obviate the binding capability of precursor cell extract in coincubation experiments. It is feasible that a transcriptional modulator present in the immature cells could be sequestered by partners present in differentiated cells, so that it is no longer available to bind to the p21 promoter. Such an interaction has been reported for Rb, which can sequester a CCAAT-binding protein required for cyclin A transcription.65 However, preclearance of our differentiated cell extracts with antibodies to Rb, E2F, p21, p107, and p53 or coincubation of extracts with T-antigen does not alter their ability to inhibit binding to the 44-bp sequence (R.S., unpublished observations).
Our finding that mature cell extracts suppress promoter element binding by extracts of immature cells is most compatible with a model of p21 regulation through active derepression. A repressor of p21 transcription is postulated to bind to the promoter within the 44-bp fragment in CD34+ cells. The repressor disengages as the cells mature, permitting p21 expression in differentiating cells. A similar mechanism of developmentally regulated activation of transcription has been reported for other genes. Activation of elastin gene expression in developing aorta,66 of the stearoyl-CoA desaturase 2 gene activation during preadipocyte differentiation,67 and of the β-globin gene during erythroid differentiation68 all involve disruption of a repressor complex. Our model proposes sequestration of a p21-promoter repressor during the normal maturation of myeloid cells as the mechanism through which derepression might occur.
ACKNOWLEDGMENT
The authors thank Xiao-Fan Wang for the p2l promoter-luciferase plasmid p21-P, David Tweardy for the SIE target fragment, Brian Dynlacht for anti-p21 antibody CP-36, Nicola Pavletich for p53 core protein, and Calbiochem for the gift of p53 Ab (MoAb-6). We are indebted to Sandra Kaplan for hematopathologic analysis; to Ying Li, Augustine Iro, and Wei Pei for excellent technical support; and to Donna Shields for CD34+ cell isolation. We are also indebted to Tim Wright, Robert Redner, Reza Zarnegar, and Qing Dou for helpful discussions of the project and manuscript.
Supported by grants from the Laurie Strauss Foundation, American Cancer Society (JRFA-594), and National Institutes of Health (HL54172-01) to R.S.
Address reprint requests to Richard A. Steinman, MD, PhD, E1052 BST, University of Pittsburgh School of Medicine, Pittsburgh, PA 15213; e-mail: Steinman+@pitt.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.