Abstract
The presence of residual leukemic cells was studied using metaphase-fluorescence in situ hybridization (FISH) in 22 patients with acute myeloid leukemia treated with chemotherapy only or chemotherapy followed by allogeneic bone marrow transplantation. The patients were followed up during their complete remission (CR) for 4 to 108 months (median, 21 months). A total of 88 BM samples was studied. In most of the samples more than 1,000 metaphase cells were analyzed. Residual leukemic cells were detected in 9 of 22 patients (41%). All patients who had an increasing and/or persisting level of abnormal cells in two or more subsequent samples or whose initial samples contained more than 1% of abnormal cells relapsed with one exception, in whom the later subsequent samples showed disappearance of abnormal cells. The time span before the first positive sample seems to be insignificant with regard to the outcome of relapse. Absence or single occurrence of abnormal cells followed by their disappearance was in agreement with CR in all the cases (16 patients). Our results indicate that metaphase-FISH is a reliable tool in the quantitation of residual leukemic cells and provides valuable prognostic information for patients with AML.
THE INITIAL chemotherapy of acute myeloid leukemia (AML) is successful in a majority of patients, and about 60% to 80% of adult patients, less than 65 years of age, enter remission as a result of induction treatment. However, despite consolidation treatment more than half of the patients relapse if treated with chemotherapy only.1,2 After allogeneic bone marrow transplantation (BMT) the risk of relapse depends on the stage of the disease at which the transplantation is performed. If the transplantation is performed in the first remission, the relapse rate is in the order of 10% to 20%, but transplantations carried out at later stages increase the risk of relapse.3,4 Cytogenetic findings have also been found to implicate the risk of relapse. Patients with t(8; 21), t(15; 17), or inv(16) have a more favorable outcome than patients with −5, −7, +8, or Philadelphia chromosome (Ph+).5 6 A method that can reliably identify in advance the patients who will suffer a relapse would be useful for planning of further treatment.
Standard cytogenetics and molecular genetic methods have been widely used for the evaluation of residual leukemic cells in clinical remission. Polymerase chain reaction (PCR) techniques can be used in AML for the detection of fusion genes originating from leukemia-specific translocations or inversions, for example, AML1/ETO [t(8; 21) (q22; q22)], PML/RARA [t(15; 17)(q22; q11-22)], and BCR/ABL [t(9; 22)(q34; q11)].7 PCR is a sensitive technique but, because only a few patients with AML have fusion transcripts, this method can only be applied to selected patients. The clinical implications of PCR+ results in different subtypes of AML patients in clinical remission are not clear. In AML-M2, for example, it has been shown that translocation t(8; 21) can be demonstrated repeatedly over several years without the disease relapsing.8-15
Unlike PCR, cytogenetic analysis allows the visualization and quantitation of clonal chromosomal aberrations, eg, translocations and trisomies. It shows that the aberrant cells are mitotic and capable of proliferating. However, the standard cytogenetic methods are time-consuming and their sensitivity is low because most often no more than 20 mitoses can be studied.
Some years ago we adapted chromosomal in situ suppression hybridization (CISS) for the follow-up of patients with hematologic neoplasms.16-18 In this study, in situ hybridization was performed on cells cultured for 12 hours in the presence of a high concentration of colcemid, which allows 1,000 or more mitotic cells to be analyzed in each sample. As in standard G-banding analysis, metaphase-fluorescence in situ hybridization (FISH) permits the visualization and quantitation of chromosomal aberrations in mitotic cells with high reliability.
PATIENTS
Among the patients treated at the Department of Medicine, Helsinki University Central Hospital, 22 patients were identified, who achieved a clinical and hematologic remission and had at diagnosis a translocation, trisomy, or some other structural chromosomal aberration suitable for follow-up with metaphase-FISH. Patients with only monosomies or small deletions were excluded from the study (Table 1). Nine patients received allogeneic BMT from an HLA-identical sibling donor. Eight patients received the transplantation in the first remission and one in the second remission (patient 20). Except for patient 20, the follow-up of the cytogenetic findings was performed in the first remission. Patients were conditioned for the transplantation with cyclophosphamide (60 mg/kg/d) for 2 days and total body irradiation 12 Gy in 6 fractions (lungs shielded not to receive more than 10 Gy). Prophylaxis against graft-versus-host disease consisted of cyclosporine for 1 year after BMT and four doses of methotrexate with or without methylprednisolone. Thirteen patients were treated with chemotherapy alone. The details of chemotherapy varied according to the protocol in use. The induction consisted of 9 to 10 days of cytarabine as continuous infusion, three doses of daunorubicin or idarubicin, and thioquanine for 9 to 10 days. The second cycle was either the same combination or a high dose of cytarabine with three doses of idarubicin. The consolidation consisted of at least two cycles, one of them a combination of mitoxantrone, etoposide, and cytarabine, and one consisting of amsacrine and a high dose of cytarabine.
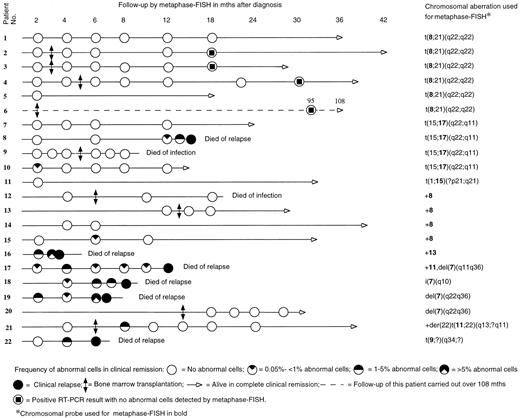
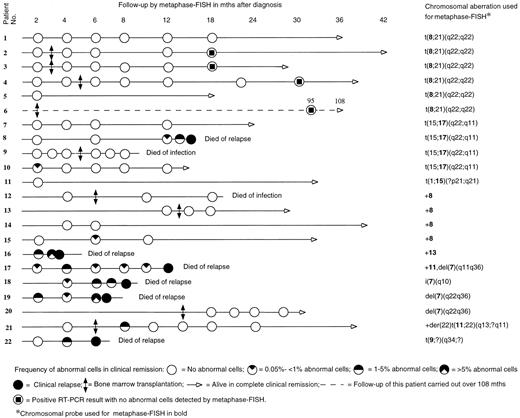
Figure 1 shows the timing of the metaphase-FISH follow-up for each patient. The duration of the clinical follow-up was 4 to 108 months, median 21 months. Altogether 88 BM samples were analyzed with metaphase-FISH. More than 1,000 cells were analyzed in 71 samples (80.7%), 500 to 1,000 in 11 samples (12.5%). Analysis of less than 500 cells was done only when abnormal cells were clearly detected (6 samples; 6.8%); hence, no more cells were required for the analysis.
Frequency of abnormal cells detected using metaphase-FISH in 22 AML patients.
MATERIALS AND METHODS
Standard cytogenetics.BM aspirates were studied at the time of diagnosis by standard cytogenetic G-banding methods following the specifications of the International Standing Committee on Human Cytogenetic Nomenclature (ISCN, 1995)21 (Table 1). At relapse, standard cytogenetic studies were also performed.
Metaphase-FISH.Standard cytogenetic preparations were used for metaphase-FISH. In this technique, only mitotic cells (mostly metaphases) were studied. Unstimulated BM cell cultures were prepared as described previously.16 Each slide contained on average 400 metaphases suitable for the analysis after in situ hybridization. About 5% of the mitoses were early metaphases, 15% mid-metaphases, 80% late metaphases and 1% to 5% were anaphases. Only clear unbroken diploid metaphase-plates with no overlapping metaphases/nuclei were analyzed. Polyploid cells were excluded and all aberrations were checked by DAPI banding to confirm the morphology and identify the chromosomal groups.
Before using a probe for follow-up, we tested it on preparations from the patient at diagnosis. Figure 1 shows the probes used for the detection of a particular chromosomal aberration. A positive metaphase cell with a translocation showed three different signals. For example, in t(8; 21) a normal chromosome 8, an abnormal chromosome 8 with unpainted material from chromosome 21, and an abnormal chromosome 21 with painted material from chromosome 8 were seen when the 8-specific probe was used, whereas in t(15; 17) a normal chromosome 17, an abnormal chromosome 17 with unpainted material from chromosome 15, and an abnormal chromosome 15 with painted material from chromosome 17 could be identified by means of the 17-specific probe. In a positive metaphase cell with trisomy 8, three copies of chromosome 8 were painted. In case of deletions, a normal chromosome and a shorter one were painted.
Chromosomal in situ suppression hybridization (CISS).CISS, also termed “chromosome painting,” was performed at diagnosis to confirm the chromosomal aberrations that were used in the follow-up. Library probes specific for chromosomes 7, 8, 9, 11, 13, 15, and 17 (American Type Culture Collection, Rockville, MD) were used for the detection of leukemia-specific translocations t(8; 21), t(15; 17), t(1; 15), and t(6; 9), trisomies, and partial deletions as described by Cremer et al.22 The probes were labeled with biotin-14-dATP by nick-translation (Nick Translation Kit; Bethesda Research Laboratories, Gaithersburg, MD). The cells were pretreated with pepsin (0.01 mg/mL; Sigma, St Louis, MO) followed by dehydration in 70%, 90%, and 96% ethanol.
For the detection of hybridization signals, avidin-conjugated fluorescein isothiocyanate (FITC; Vector Laboratories Inc, Burlingame, CA) was used.23 The cells were counterstained with 4′,6-diamidino-2-phenylindole-dihydrochloride (DAPI; Sigma) and propidium iodide (Sigma).
The cells were mounted with VECTASHIELD (Vector) antifading solution and analyzed using a Zeiss (Oberkochen, Germany) fluorescence photomicroscope with Zeiss filters 02 (FITC) and 09 (DAPI). In most of the cases, 1,000 to 2,000 metaphases per sample were available for the analysis when two to four slides were scanned. The details of follow-up and survival are presented in Fig 1.
Controls.BM aspirates from ten healthy BM donors were used as controls. In situ hybridization was performed with the same probes used in the follow-up of the patients to count the frequency of chromosomal aberrations identified in our patients and/or false-positive signals among our controls. At least 1,000 metaphases were analyzed for each probe in each sample. Because detection of isochromosomes, interstitial deletions, and insertions could be biased by the quality of metaphases, control slides were analyzed blindly against slides from patients with del(7)(q11q36), i(7)(q10), or t(9; ?)(q34; ?). Additionally, 10 AML patients without t(8; 21), t(15; 17), or +8 were tested for the frequency of false-positive signals using library probes specific for chromosomes 8 and 17 in the analysis of 1,000 metaphases from each of them. The same criteria for the interpretation were applied as above.
RESULTS
At diagnosis, leukemia-specific translocations t(8; 21) and t(15; 17) or variant t(15; 17) “der(1)t(1; 15)(?p21; q21),der(17)” were detected in patients 1 through 11, while other aberrations, +8, +11, +13, del(7), t(11; 22), and t(9; ?) were seen in the remaining patients (Table 1).
Controls.None of our 10 controls showed single cells with t(8; 21), t(15; 17), t(11; 22), or t(9; ?). For trisomy 8, only 1 metaphase in 10,000 cells (0.01%) showed an additional chromosome 8. For this metaphase DAPI banding showed a chromosome number of 47 and confirmed an extra chromosome 8. Other chromosomal aberrations, +11, +13, del(7) (q11q36), del(7)(q22q36), or i(7)(q10), were not detected in any of the 10 control specimens.
Metaphase-FISH.In 13 patients (patients 1 through 7, 9, 11 through 14, 20), no abnormal cells were detected in any of the tested samples, and all of them continued in remission at the end of the follow-up. Nine patients had abnormal cells at one or more times during the follow-up (patients 8, 10, 15 through 19, 21, 22). Absence or single occurrence of abnormal cells followed by their disappearance was in agreement with complete remission (CR) in all the cases (16 patients). The time of the first appearance ranged from 2 to 8 months. There was a marked difference between patients with different cytogenetic findings in relation to the detection of abnormal cells. Three of 15 patients (20%) with t(8; 21), t(15; 17), variant t(15; 17), or trisomy 8 exhibited abnormal cells, whereas 6 of 7 patients (85.7%) with other cytogenetic abnormalities had abnormal cells. The details of follow-up samples are presented in Fig 1.
All patients who had an increasing and/or persisting level of abnormal cells in two or more subsequent samples or whose initial samples contained more than 1% of abnormal cells relapsed, with one exception: patient 21, who remains in CR. Shortly after the allogeneic BMT the frequency of abnormal cells in patient 21 was 1.6%, but no abnormal cells were detected later in the four subsequent samples.
In five patients (8, 10, 15, 17, and 18) the frequency of abnormal cells was less than 1% (0.05% to 0.8%) in the first positive sample. Two of them (patients 10 and 15) had no further positive samples and they remain in CR, whereas the other three (patients 8, 17, and 18) showed an increasing and/or persisting level of abnormal cells in subsequent samples, and they relapsed. The time span before the first positive sample seems to be insignificant with regard to the outcome of relapse.
None of the six patients with t(8; 21) showed abnormal cells in any of the metaphase-FISH examination(s). Four of these patients had received BMT. Two of the five patients with t(15; 17) or variant t(15; 17), both treated with chemotherapy alone, had abnormal cells. One of them (patient 8) had a relatively late appearance, at 12 months, of abnormal cells which increased in a subsequent sample, and he relapsed. The other (patient 10) showed abnormal cells shortly after the treatment but these disappeared thereafter, and she remains in CR. The only transplanted patient with t(15; 17) had no abnormal cells at any time and remains in CR. Of the four patients with trisomy 8, two had been transplanted. One nontransplanted patient (no. 15) showed once a low number of abnormal cells (0.05%), which disappeared in a subsequent sample, and she remains in CR. Four patients had structural abnormalities of chromosome 7 and three of them were treated with chemotherapy while one received BMT. The transplanted patient did not show abnormal cells in any samples and remains in CR. The other three patients had an increasing and/or persisting level of abnormal cells in subsequent samples, and all three relapsed.
RT-PCR.The four transplanted patients with t(8; 21) were tested during CR with RT-PCR at time intervals shown in Fig 1. All samples, which were tested and found negative by metaphase-FISH, were positive with RT-PCR. The four patients have remained in CR for 18 to 108 months. The RT-PCR results have been published elsewhere.15
DISCUSSION
In this study, a clonal chromosomal aberration detected at diagnosis was selected for follow-up using metaphase-FISH. Standard chromosome banding analysis is usually based on a selection of 20 good-quality metaphases which rules out approximately 14 × 10−2 mosaicism at a 0.95 confidence limit based on the statistical formula provided by Hook (1977).24 On the other hand, metaphase-FISH allows analysis of as many as 2,000 mitotic cells which need not be selected. This technique also enables the demonstration of an aberration at chromosome banding level. Thus, metaphase-FISH is at least as reliable as chromosome banding analysis and, importantly, it ruled out mosaicism present at 14 × 10−4 on a 0.95 confidence limit, when we applied the same statistical formula.
Using metaphase-FISH we found leukemic cells in 9 of 22 patients (41%) in CR. The occurrence of abnormal cells was rare in patients with t(8; 21), t(15; 17), or trisomy 8, but it was more common in patients with other aberrations, the majority of them having structural abnormalities of chromosome 7. Therefore, variations in the presence of residual leukemic cells were dependent on the initial cytogenetic finding and represented the differences in the known prognostic groups where patients with t(8; 21) or t(15; 17) have a good prognosis, trisomy 8 of an intermediate prognosis and del(7) of a poor outcome.5 6
The possibility to monitor the behavior of low numbers of residual leukemic cells is important for the understanding of the biology of leukemia. A crucial question is the impact of metaphase-FISH findings on clinical decision-making. When effective treatment alternatives, such as BMT, are available, it is clinically important to recognize in advance the patients who will suffer a relapse and to give the treatment when the leukemic cell burden is low. We found that the presence of low numbers of residual leukemic cells (<1%) does not necessarily predict an imminent relapse, because these cells may disappear in subsequent samples. Instead, patients who show abnormal cells at frequencies higher than 1% or present repeated positive samples (increasing or persisting level of abnormal cells) are at a very high risk to relapse and need further treatment. The timing of the first detection of low numbers of leukemic cells in a single sample was not helpful in the prediction of outcome. In this respect our results correspond to our previous results of CML patients followed up using metaphase-FISH after allogeneic BMT.16
The implications of the demonstration of residual leukemic cells may not be similar after different treatments. After allogeneic BMT, graft-versus-leukemia effect may modify the behavior of the leukemic clone differing from the situation after chemotherapy only. Graft-versus-leukemia effect is an important part of the curative impact of allogeneic transplantation. This effect is most marked in CML and its presence has also been clearly demonstrated in other myeloid malignancies.25-27 Patient 21 in this material may show this effect. He had a significant number of residual leukemic cells (1.6%) in the BM shortly after allogeneic BMT, but further samples were negative. It should be noted that among those patients who had an abnormality other than t(8; 21), t(15; 17), or trisomy 8, only the two allogeneic BMT recipients (patients 20 and 21) had mostly negative metaphase-FISH samples and a good outcome. None of the transplanted patients relapsed in this study. Therefore, the demonstration of residual leukemic cells shortly after allogeneic BMT is likely to impact the prognosis different from a similar finding after chemotherapy.
As previous studies by RT-PCR have shown that the AML1/ETO fusion transcript of t(8; 21) may be present in remission patients for years after BMT,8-15 the RT-PCR study was also performed on four patients with t(8; 21), whose remission had lasted for 2 to 9 years.15 The transcript was found in all of these patients, but no abnormal cells were seen by metaphase-FISH. PCR is a highly sensitive technique and the biologic significance of its positive results in remission patients remains to be determined. However, in patients with t(15; 17), PCR results have correlated positively with the clinical outcome, where the presence of abnormal cells has been a sign of impending relapse.28-30
This study shows that metaphase-FISH is a useful, sensitive, and reliable technique for the demonstration of residual leukemic cells in patients with AML in CR, whose clonal chromosome aberration is a trisomy, deletion, or a translocation. Visualization of both chromosomal morphology and hybridization signals in abnormal metaphases eliminates the problem of false-positive cells, typical to interphase-FISH where hybridization signals are used as markers of aberrations. Furthermore, metaphase-FISH is a quantitative technique that analyzes a large number of cells, up to 2,000 metaphases per sample, a feature which helps to evaluate the clinical significance of the finding and is useful in planning further treatment.
Supported by the Finnish Cancer Society.
Address reprint requests to Sakari Knuutila, PhD, Department of Medical Genetics, Haartman Institute, PO Box 21 (Haartmaninkatu 3), FIN-00014 University of Helsinki, Finland.