Introduction: Human cytomegalovirus (HCMV) is a b-herpesvirus that is highly prevalent in the adult population and has the ability to establish lifelong latency in healthy individuals. The innate and adaptive immune systems work closely to control viral replication, leading to a dynamic interaction between the virus and the host immune system. This interplay leads to the development of distinct immune-cell repertoires in HCMV seropositive (HCMV +) individuals. Natural killer (NK) cells are cytotoxic innate lymphocytes that are required to manage and control HCMV infections. NK cells utilize their NKG2C/CD94 receptor complex to mount a response to infected cells that present HLA-E molecules loaded with the HCMV UL40-derived peptide. As a result, HCMV + individuals possess higher levels of NKG2C + NK cells, and within this population, there is a distinct subset referred to as “memory” due to their adaptive-like characteristics. The developmental origins and molecular mechanisms involved in the maintenance and persistence of these cells remain largely unknown. In our current study, we investigate the origins and transcriptional signatures of persistent memory NKG2C + NK cells obtained from human donor spleens.
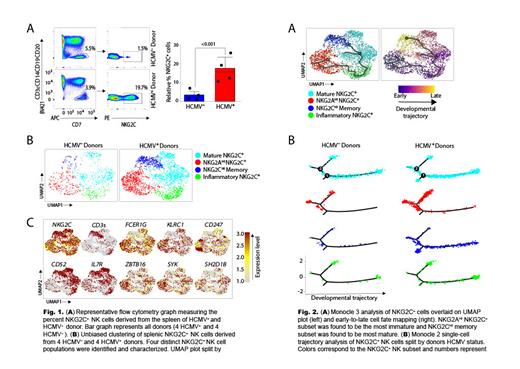
Methods & Results: Eight healthy adult human spleens were obtained from four HCMV + and four HCMV seronegative (HCMV -) donors. Donor median age was 59 [IQR 48.5-56.5], 50% (n=4) were identified as female, 50% (n=2) of females were HCMV + and 50% (n=2) of females were HCMV -. Spleens were provided by the Versiti Organ Donor Center of Wisconsin and were processed to a single cell suspension. In line with previous findings in the peripheral blood of HCMV + individuals, we observed significantly elevated levels of NKG2C + NK cells in the spleens of HCMV + donors ( Fig. 1A). This observed significance of higher NKG2C + NK cells was consistent across all HCMV + donors when compared to our HCMV - donors ( Fig. 1A).
To investigate the molecular mechanisms involved in the maintenance and persistence of memory NKG2C + NK cells, we performed single-cell RNA sequencing (scRNA-seq), using the sorted NK cells from all eight donors. Using unbiased clustering analysis, we identified and characterized four distinct NKG2C + splenic NK cell subsets ( Fig. 1B). Our findings indicated that the relative composition of these subsets was highly influenced by the HCMV status of the donors. Specifically, we observed that HCMV + donors had significantly higher levels of NKG2C Hi memory NK cells ( Fig. 1B). This NKG2C Hi memory NK subset had significantly higher expression of NKG2C, CD52, CD3 e, and IL7R ( Fig. 1C) and significantly lower expression of FCER1G, KLRC1, CD247, ZBTB16, SYK, and SH2D1B ( Fig. 1C). These findings suggest NKG2C Hi memory NK cells possess unique transcriptional and molecular mechanisms that may contribute to their ability to persist over time.
To explore the developmental cell fate of NKG2C Hi memory NK, we utilized both the Monocle 2 and Monocle 3 software's to track how NKG2C + cells transition between transcriptomic states. Monocle 3 analysis yielded a simple early-to-late cell fate trajectory that was projected onto our UMAP plot ( Fig. 2A). We found that NKG2A Hi NKG2C + subset served as the early timepoint in the developmental trajectory, ultimately leading to the development of NKG2C Hi memory NK ( Fig. 2A). The Monocle 2 analysis produced unique developmental trajectory plots, allowing us to visualize distinct developmental pathways and transitions ( Fig. 2B). Interestingly, we observed a unique branch-point exclusive to the HCMV + donors ( Fig. 2B) that expressed significantly higher levels of CD3 e and IL7R. These observations could indicate a specific developmental pathway unique to NKG2C Hi memory NK.
Conclusion: Here, we demonstrate that HCMV infection can induce the formation of NKG2C Hi memory NK cell subset that displays a unique transcriptional and developmental profile. These findings can influence the future isolation and application of memory NK cells in cellular immunotherapies.
Disclosures
No relevant conflicts of interest to declare.