Abstract
Background: Immunotherapeutics targeting B cell maturation antigen (BCMA), including belantamab mafodotin (BelaMaf), idecabtagene vicleucel, and ciltacabtagene autoleucel, have revolutionized treatment in relapsed and/or refractory myeloma (RRMM). However, our understanding of how these therapies change both the molecular and immune characteristics of the myeloma clones and their tumor microenvironment (TME) is incomplete and hinders our ability to rationally sequence therapies.
Methods: We performed single-cell RNA sequencing (scRNA-seq) to comprehensively characterize the cellular and molecular properties of myeloma cells and the immunological characteristics of TME to establish the landscape and dynamics of the tumor-immune ecosystem in RRMM treated with BCMA-based immunotherapies. From April, 2021, until January, 2022, 16 patients were eligible for whom scRNA-seq data have been finalized. Baseline bone marrow (BM) aspirates were obtained and patients were classified according to their response to BCMA targeted therapy. This work was supported by Dr. Miriam and Sheldon G. Adelson Medical Research Foundation and the Paula and Rodger Riney Foundation's Riney Family Multiple Myeloma Research Fund at MD Anderson.
Results: At the time of BM collection, seven patients were BCMA therapy-naïve with all seven later achieving a response to BCMA-targeted therapy whereas nine patients were BCMA-refractory. Fresh aspirates were processed through CD138+ selection (n=10) or processed as whole BM (WBM) (n=6). cDNA libraries from enriched CD138+ (n=10) and CD138- cell compartments (n=10) as well as WBM samples (n=6), were then used for simultaneous 5’ gene expression (scRNAseq) and T/B cell receptor sequencing (scTCR/BCRseq). We profiled 104,638 high-quality cells including 56,803 CD138+ cells and 47,835 TME cells. We obtained scTCRseq and scBCRseq data on 14,553 and 62,215 cells, respectively. Among them, 58,703 out of 76,768 cells had paired scRNA-seq data.
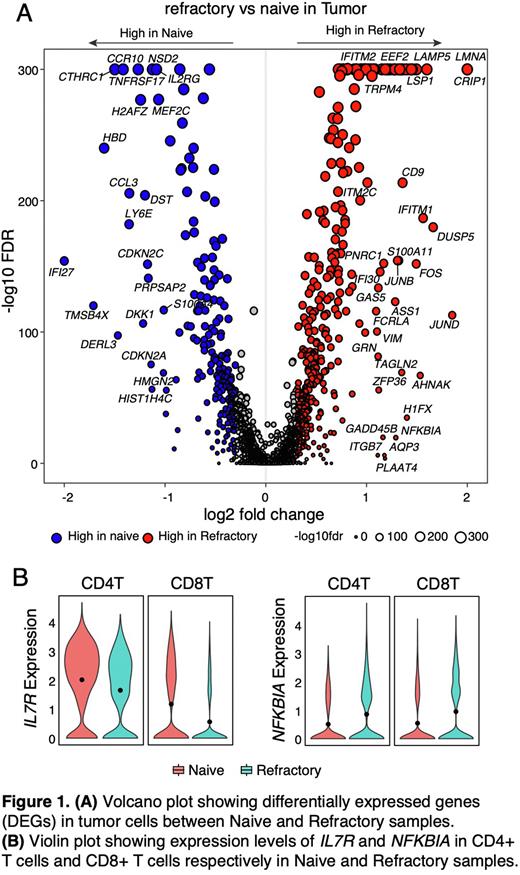
Unsupervised clustering analysis revealed 13 different TME cell types and various CD138+ cell states. Malignant cells were identified based on inferred copy number alterations. Tumor and TME cells from naïve and refractory samples clustered distinctly, uncovering unique transcriptomic profiles. Differential gene expression analysis in non-refractory vs refractory patients’ tumor cells showed high expression of BCMA, CTHRC1, CCR10, NSD2 and IL2RG in non-refractory vs high expression of EEF2, IFITM2, LMNA, CRIP1, LAMP5 and TRPM4 in refractory samples. Pathway analysis revealed upregulated activity of TNFα via NF-kB, P53, MYC and hypoxia pathways and downregulated Interferon α and γ response pathways in refractory compared to non-refractory patients (Figure 1A). We next evaluated the TME cell compositions of refractory vs. non-refractory patients and observed a trend of increased myeloid cells (CD14+ and CD16+ monocytes) and fibroblasts and decreased conventional dendritic (cDC2) and NK cells which could contribute to creating an immunosuppressive TME. Further subclustering analysis of T cells identified 10 CD4 and 8 CD8 T cell states, and we observed a trend of increased GZMK+CXCR3+TNFAIP3+ CD4 T cell state and decreased activated, and cytotoxic CD8 T cell states in refractory when compared to naïve patients. We also noted that the IL-7 receptor that regulates key signaling of T cell survival was significantly decreased and NFKB1 that encodes the core component of the canonical NF-κB pathway was significantly increased in CD4 and CD8 T cells from refractory (vs. naïve) patients (Figure 1B).
Conclusions: Overall, BCMA-refractory patients have a TME landscape consistent with a more suppressed and inactive immune microenvironment than non-refractory patients. Upregulation of key oncogene and tumor suppressor gene pathways including p53, MYC, and NF-κB underlie tumor refractoriness, suggesting that novel therapies with different mechanisms of action are needed. High tumor expression of LAMP5 in BCMA refractory patients suggests the potential for LAMP5 targeting. Using BCMA-targeting therapies earlier in the disease course, with a more robust immune microenvironment, may increase and deepen response to treatment. Updated data will be presented at the Meeting on a total cohort of 35 patients.
Disclosures
Lee:Regeneron: Research Funding; Oncopetides: Consultancy; Karyopharm: Consultancy; Takeda Pharmaceuticals: Consultancy, Research Funding; GSK: Consultancy, Research Funding; Amgen: Research Funding; Monte Rosa Therapeutics: Consultancy; Sanofi: Consultancy; Genentech: Consultancy; Immunitas Therapeutics: Consultancy; Legend Biotech: Consultancy; Pfizer: Consultancy; Janssen: Research Funding. Patel:Janssen, Celgene/BMS, Caribou Sciences, Arcellx, Cellectis, Merck, Pfizer, Karyopharm, Oncopeptides: Consultancy. Thomas:Genentech: Research Funding; Bristol Myers Squibb: Research Funding; Janssen Pharma: Research Funding; X4 Pharma: Research Funding; Cellectar Pharma: Research Funding; Ascentage Pharma: Research Funding. Orlowski:Abbvie, BioTheryX, Inc., Bristol-Myers Squibb, Janssen Biotech, Karyopharm Therapeutics, Inc., Meridian Therapeutics, Monte Rosa Therapeutics, Neoleukin Corporation, Oncopeptides AB, Regeneron Pharmaceuticals, Inc., Sanofi-Aventis, and Takeda Pharmaceutic: Honoraria, Membership on an entity's Board of Directors or advisory committees; Asylia Therapeutics, Inc.: Current equity holder in private company; CARsgen Therapeutics, Celgene/Bristol Myers Squibb, Exelixis, Janssen Biotech, Sanofi-Aventis, Takeda Pharmaceuticals North America, Inc.: Research Funding; Asylia Therapeutics, Inc., BioTheryX, Inc., Heidelberg Pharma, Inc.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.