Abstract
Background: Hematologic malignancies are a spectrum of disease that frequently have an insidious onset and may be difficult to diagnose. Improved screening using basic complete blood count parameters are needed to enhance early detection of these processes. Aging results in an expansion of hematopoietic clones that may predispose individuals to subsequent development of a hematologic malignancy. Recent studies with next generation sequencing (NGS) and whole-exome sequencing have identified certain somatic mutations known as DTA mutations that are associated with aging (e.g., DNMT3A, TET2, and ASXL1). These mutations, although more commonly seen in myeloid malignancies, are initially benign but may develop into overt hematologic malignancy and are collectively termed clonal hematopoiesis of indeterminate potential (CHIP). Similarly, individuals with cytopenia and clonal mutations are designated as clonal cytopenia of undetermined significance (CCUS). However, these mutations carry a risk of progression to hematologic malignancy. Animal models have shown that clonal hematopoiesis can accelerate cardiovascular mortality due to increased atherogenesis by causing inflammation and endothelial injury via upregulation of inflammasome pathways. Also, it has already been shown that clonal hematopoiesis with increasing number of clonal mutations, higher variant allele frequency and mutations in known driver genes have a higher risk of progression to hematopoietic malignancy (acute myeloid leukemia, AML). Certain acute phase reactants, like C-reactive protein (CRP) or erythrocyte sedimentation rate (ESR) have been used in prior studies as markers of inflammation and are strongly associated with variations in red cell size as determined by the red cell distribution width (RDW), reflecting chimeric clonal origins. We therefore propose that RDW and mean corpuscular volume (MCV) with or without other blood count parameters (red cell count, white blood cell count and platelet count) might be used as important predictors of progression of myeloid neoplasms, especially myelodysplastic syndromes (MDS), clonal cytopenias (CCUS) and acute myeloid leukemias (AML).
Methods: Utilizing Epic's self-service reporting tool SlicerDicer, cohort discovery was performed on the Caboodle Patients data model. Cohort criteria included individuals with the cytopenia diagnostic grouper that had undergone blood or bone marrow myeloid panel next generation sequencing, that also had concurrent or historic complete blood count (CBC) results. Patients with known acute leukemias were excluded. Once the cohort was identified, a data pull was performed that captured CBC values, age, gender, race, and ethnicity, in addition to the NGS results. The NGS results were ranked into 3 classes from 0 to 2. Class 0 included results with no apparent disease, while class 2 results included high grade myeloid mutations (i.e. mutations in NRAS, KRAS, TP53, RUNX1 or ASKL1). Class 1 mutations were considered low grade and featured mutations not seen in class 2. The collated data was analyzed using SAS software.
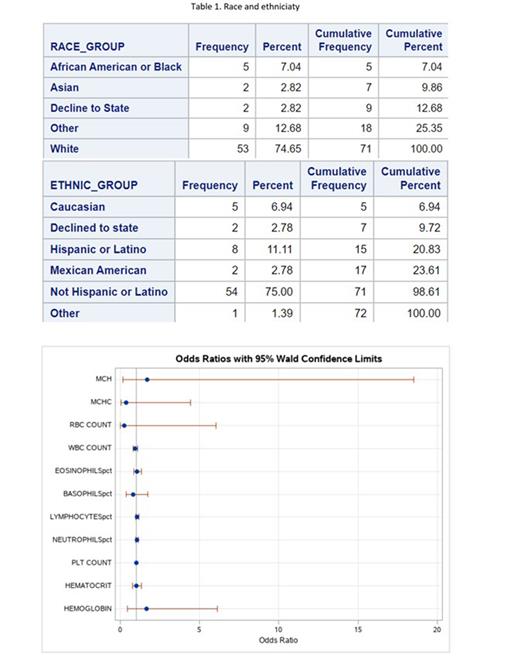
Results: Overall 72 patients met inclusion criteria. Patients ranged in age from 23 to 92 years, 48% identified as female and 52% identified as male with a mixture of race and ethnicity (Table 1). Abnormalities in several CBC components including MCH, MCHC, RBC count, and hemoglobin concentration were associated with increased odds of identifying those with malignant myeloid processes (Figure 1). It is notable that no mutations in FLT3 or KIT were observed in this cohort.
Discussion: Based on our preliminary data, these cost-effective parameters can be added to an algorithm for risk assessment of hematologic neoplasms and better predict which patients will further benefit from targeted gene testing. Also, additional data will be helpful and applying this data to modern machine learning techniques will further the clinical application of these findings.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.