Abstract
Background: In order to gain a deeper understanding of the clinical, molecular and immune parameters involved in multiple myeloma (MM) disease initiation, progression and response to treatment, the Multiple Myleoma Research Foundation (MMRF) and its partners launched the CureCloud Research Initiative (NCT03657251), a Direct-to-Patient (DTP) effort aimed at enrolling 5,000 individuals from whom comprehensive molecular and immune analyses are generated from blood specimens and the resulting data aggregated with the correlating clinical information.
Methods: To support the molecular characterization of liquid biopsies for CureCloud, a set of myeloma-specific blood-based approaches were developed. The first of such assays is a hybrid selection panel that captures 70 commonly altered MM and Clonal Hematopoiesis of Indeterminate Potential (CHIP) genes detecting somatic variants present in a patient's circulating-free DNA (cfDNA). This MM-70 assay was thoroughly validated to >90% sensitivity for events at 1% variant allele frequency with a specificity of <0.2 FP/Mb when sequenced at 60,000x. A clinical-grade (CLIA) pipeline was established enabling the return of results. Several quality control measures were used to select for CHIP related mutations including filtering out known myeloma driver events, removing high frequency events, and overlapping with previously published CHIP related mutations.
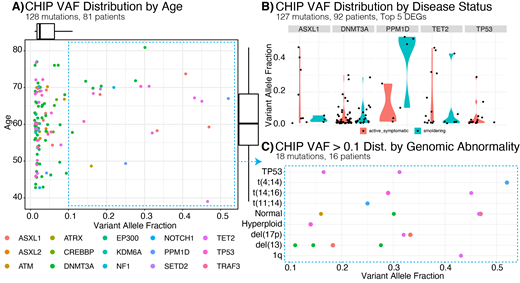
Results: 580 subjects were enrolled to the study at abstract submission, including 127 smolderers, 173 newly diagnosed and 265 relapsed participants. 430 cases have been analyzed on the MM-70 assay. A mean cfDNA amount of 31 ng was recovered for input. Mean raw coverage depth of 176,000x was attained, with a mean duplex depth of 1271 (517-2406). 417 cases passed the minimum CLIA validation thresholds. Some of the most frequently mutated genes observed were in the MAPK, DNA repair, epigenetic, cell cycle, and NF-kappaB pathways in agreement with MM retrospective genomic research studies performed on bone marrow aspirates. Of interest, CHIP mutations were detected in a large proportion of cases (174) with their prevalence increasing with age (Figure 1A). 117 of patients had only one CHIP mutation while 57 had two or more with the same genomic loci were recurrently mutated in DNMT3A, TP53, ASXL1, and PPM1D. No significant differences in CHIP were observed between smoldering and overt myeloma subjects (Figure 1B). Interestingly, a greater (>0.1) CHIP Variant Allelic Fraction (VAF) was seen in individuals with higher risk genomic alterations (Figure 1C).
Conclusion: This genomic characterization of one of the largest cohorts to date of individuals with myeloma and precursor conditions using a cfDNA-based liquid biopsy assay was able to recapitulate known myeloma-specific mutations and reinforced the importance of CHIP alterations in MM. CHIP mutations are indeed frequent in myeloma, increase with age, and higher burden might be related to disease aggressiveness.
Ghobrial: AbbVie, Adaptive, Aptitude Health, BMS, Cellectar, Curio Science, Genetch, Janssen, Janssen Central American and Caribbean, Karyopharm, Medscape, Oncopeptides, Sanofi, Takeda, The Binding Site, GNS, GSK: Consultancy.