In this issue of Blood,Wagener et al demonstrate a new World Health Organization (WHO) provisional entity of MYC– Burkitt-like lymphoma (BLL) with 11q aberrations (mnBLL, 11q,) that is distinct from MYC+ sporadic Burkitt lymphoma (BL) as well as other germinal center–derived B-cell lymphomas, including follicular lymphoma and diffuse large B-cell lymphoma.1
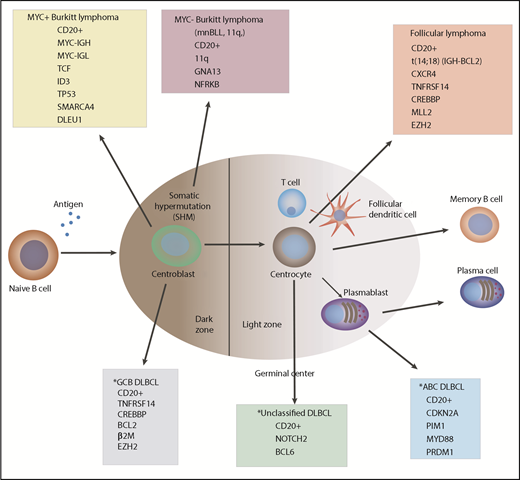
Germinal center–derived B-cell lymphomagenesis. *Diffuse large B-cell lymphoma (DLBCL), recently subclassified as MCD, BNS, N1, and EZB subgroups.10
Germinal center–derived B-cell lymphomagenesis. *Diffuse large B-cell lymphoma (DLBCL), recently subclassified as MCD, BNS, N1, and EZB subgroups.10
As demonstrated by array-based imbalance mapping and whole-exome sequencing, these significant findings provide evidence of the genetic signature associated with this new subtype of MYC– BL. Furthermore, Wagener et al demonstrate potential therapeutic target genes of this MYC– BL, suggesting new methods that could be used in the differential diagnostic process of distinguishing MYC+ sporadic BL from mnBLL, 11q, lymphomas.
Our group initially reported the presence of derivative chromosome 11q (der11q) using standard cytogenetic karyotyping in a small number of children with BLL (on the basis of the Revised European-American Classification of Lymphoid Neoplasms [REAL] Classification) who were treated on the Children’s Cancer Group 5961 international French-American-British mature B-NHL (LMB) trial.2-4 Subsequently, Salaverria et al5 reported on a new pathologic entity of MYC– high-grade B-cell lymphoma similar to BL in morphology but characterized by a recurrent 11q aberration pattern by single nucleotide polymorphism and comparative genomic hybridization arrays. In the WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues (5th edition), a new provisional diagnosis was made for B-cell lymphomas that resembled BL morphologically and immunophenotypically and by gene expression profiling, except they lack MYC gene rearrangements or amplified expression and are characterized by 11q aberrations.6
Wagener et al now provide new molecular evidence by array-based imbalance mapping and WES of a distinct genetic signature of this new pathological entity of mnBLL, 11q,. This signature is unique when compared with MYC+ sporadic BL, FLL, and germinal center B-cell (GCB)–derived DLBCL. The genetic mutational landscape of mnBLL, 11q, is characterized by GNA13 mutations (47%), NFRKB mutations (27%), and INO80 chromatin remodeling complex mutations (33%). These results suggest a role of the GNA13 gene in the pathogenesis of mnBLL, 11q, and the NFRKB gene as part of the INO80 complex as a potential candidate gene in the deleted region of 11q24.3. Importantly, the latter gene complex (INO80) has been reported to play an important role in transcriptional regulation.
Additional chromosomal imbalances were identified in MYC+ BLL that were located outside chromosome 11. These other cytogenetic abnormalities include partial trisomy 12 (47%) (12q13.11-q24.32), gains in 7q (+7q), and loss in the 13q32.3-q34 region. The same cytogenetic abnormalities were identified by routine chromosomal karyotyping almost 10 years ago in children with BLL defined by the REAL Classification.4 Furthermore, in the Wagener et al study, genes commonly altered in expression and/or mutated in MYC+ sporadic BL, such as ID3, TCF3, TP53, DLEU1, SMARCA4, and the SWI/SNF complex, were not recurrently (>15%) mutated in patients with mnBLL, 11q, (see figure).7,8 DLEU1 located in the region of 13q14.3 in MYC+ sporadic BL has been identified as a potential tumor suppressor gene and confers chemoimmunotherapy resistance in MYC+ BL.9 Further research will be required to determine the clinical significance of these new mutations associated with mnBLL, 11q, and what role, if any, these mutations play in the pathogenesis and/or sensitivity or resistance in the treatment of this rare B-cell lymphoma.
In summary, the findings by Wagener et al significantly add to our knowledge base regarding the molecular heterogeneity of germinal center–derived B-cell lymphomas in both children and adults (see figure).10 Further elucidation of the molecular basis of the pathogenesis and/or resistance of all B-cell lymphomas will provide opportunities to investigate new therapeutic strategies for treating these malignant disorders.
Conflict-of-interest disclosure: The author declares no competing financial interests.