In this issue of Blood, Chinn et al report genetic findings in a large cohort of children with hemophagocytic lymphohistiocytosis (HLH), indicating that many cases may be explained by mutations in genes other than those required for lymphocyte cytotoxicity.1
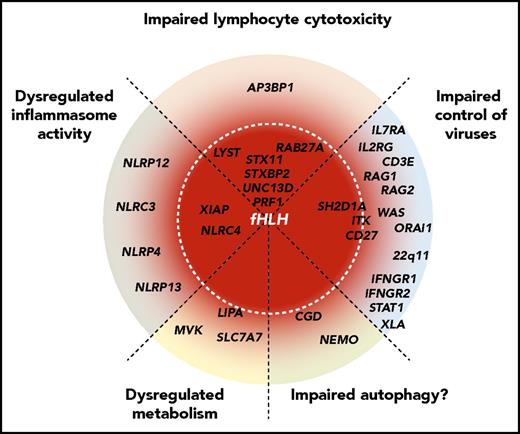
Schematic representation of the spectrum of HLH-predisposing genetic conditions, representing different pathogenic mechanisms. Genes are grouped by pathogenic mechanisms and radially organized by their associated risk of HLH, where genes at the center are fully penetrant. Mutations in genes required for lymphocyte cytotoxicity invariably lead to HLH in infancy. Mutations in other genes that impair control of common viruses via impaired lymphocyte signaling (eg, SH2D1A,ITK, and CD27) are often associated with HLH, whereas impaired lymphocyte development and function (genes causative of severe combined immunodeficiency or combined immunodeficiency) may also predispose patients to HLH. Moreover, patients with more general defects in interferon signaling (eg, genes causative of Mendelian susceptibility to mycobacterial diseases) may develop HLH. Constitutive activation or dysregulation of the inflammasome, as observed in patients with activating NLRC4 mutations or loss-of-function XIAP mutations, can cause HLH through primary macrophage activation. This might also be the case for genetic variants in other DIAP genes, as revealed by Chinn et al. Similarly, even in metabolic diseases such as Wolman disease (LIPA), primary macrophage activation resulting from accumulation of metabolites might constitute the initial trigger for HLH development. In chronic granulomatous disease, impaired autophagy may represent a mechanism for HLH susceptibility.
Schematic representation of the spectrum of HLH-predisposing genetic conditions, representing different pathogenic mechanisms. Genes are grouped by pathogenic mechanisms and radially organized by their associated risk of HLH, where genes at the center are fully penetrant. Mutations in genes required for lymphocyte cytotoxicity invariably lead to HLH in infancy. Mutations in other genes that impair control of common viruses via impaired lymphocyte signaling (eg, SH2D1A,ITK, and CD27) are often associated with HLH, whereas impaired lymphocyte development and function (genes causative of severe combined immunodeficiency or combined immunodeficiency) may also predispose patients to HLH. Moreover, patients with more general defects in interferon signaling (eg, genes causative of Mendelian susceptibility to mycobacterial diseases) may develop HLH. Constitutive activation or dysregulation of the inflammasome, as observed in patients with activating NLRC4 mutations or loss-of-function XIAP mutations, can cause HLH through primary macrophage activation. This might also be the case for genetic variants in other DIAP genes, as revealed by Chinn et al. Similarly, even in metabolic diseases such as Wolman disease (LIPA), primary macrophage activation resulting from accumulation of metabolites might constitute the initial trigger for HLH development. In chronic granulomatous disease, impaired autophagy may represent a mechanism for HLH susceptibility.
HLH is a life-threatening hyperinflammatory syndrome characterized by unremitting fever, cytopenia, and hepatosplenomegaly, which together represent signs of dysregulated immunological homeostasis. Clinically, HLH is diagnosed upon fulfillment of 5 of 8 clinical signs and laboratory parameters, commonly referred to as the HLH-2004 diagnostic criteria.2 Familial HLH (fHLH) has been associated with autosomal recessive mutations in genes required for lymphocyte cytotoxicity (ie, PRF1,UNC13D,STX11,STXBP2,RAB27A, and LYST).3 A hallmark of these forms of primary HLH, caused by fHLH-associated mutations, is impaired lymphocyte cytotoxicity, which also constitutes 1 of the HLH-2004 diagnostic criteria. Moreover, mutations in SH2D1A and XIAP cause X-linked forms of fHLH, which exhibit distinct cellular mechanisms. These genes encode proteins involved in lymphocyte signaling as well as regulation of apoptosis and inflammasome activity, respectively.3 Hematopoietic stem-cell transplantation represents the only curative treatment of primary HLH. However, if defects in lymphocyte cytotoxicity are not present, a less aggressive treatment strategy may be appropriate. Patients lacking fHLH-associated mutations are often described as having secondary HLH. Underlying conditions such as infections, rheumatism, or malignant diseases represent a common feature of secondary HLH, the pathogenesis of which is less well understood. Notably, patients with secondary HLH may harbor other genetic predispositions.3 For example, metabolic disorders and other primary immunodeficiency diseases (PIDs) occasionally present with HLH, thus falling under the umbrella of secondary HLH.4,5 However, the degree to which genetic factors predispose patients to secondary HLH has not been comprehensively determined.
Genetic testing for fHLH has been available since 1999, when mutations in PRF1 were first discovered. Patients have typically been evaluated by a sequential gene sequencing approach, which is labor intensive and time consuming. Targeted sequencing has also been guided by the results of natural killer cell cytotoxicity and exocytosis, achieving the highest diagnostic yields in patients with proven defective lymphocyte cytotoxicity.6,7 The introduction of high-throughput sequencing in research as well as clinical diagnostics during the last 10 years has radically changed the molecular approach to rare pediatric diseases, including HLH and the broader group of PIDs.8 Comprehensive sequencing efforts promise a fuller understanding of HLH predisposition and pathogenesis, enabling precision medicine.
Encompassing >17 years of experience in diagnosing and managing HLH at a large tertiary care center, Chinn et al report their findings in a prospective multiethnic cohort of 122 children fulfilling the HLH-2004 criteria. A vast majority of these children underwent genetic testing for fHLH. Moreover, exome sequencing was performed in 48, representing the largest exome data set hitherto published for HLH. A definitive molecular diagnosis of fHLH was only achieved in a limited number of patients (n = 19; 19%), with fHLH overrepresented in infants diagnosed before 1 year of age (61%). By exome sequencing, a group of patients were diagnosed with other well-known PIDs, such as Omenn syndrome, chronic granulomatous disease, and autoimmune lymphoproliferative syndrome, corroborating previous findings from Bode et al.4 Taking advantage of a large cohort of individuals undergoing sequencing with the exome platform at the Baylor-Hopkins Center for Mendelian Genomics, the authors were also able to test for enrichment of digenic inheritance in patients fulfilling HLH-2004 criteria compared with nearly 6000 control individuals. In their data, they did not find statistical support for a digenic model of susceptibility to HLH. Instead, they uncovered an association between HLH and genetic variants in a group of genes defined by the authors as dysregulated immune activation or proliferation (DIAP) genes, including significant associations for monoallelic variants in NLRC4 and NLRP12 and biallelic variants in NLRP4, NLRC3, and NLRP13. Although heterozygous activating mutations in NLRC4 have previously been shown to cause severe HLH and macrophage activation syndrome,9 the associations between HLH and other inflammasome genes are novel. A stratification of clinical features, such as age at onset, survival, and presence of known HLH-associated triggers based on genetic findings, revealed that isolated DIAP-associated variants were in most cases associated with somewhat milder rheumatic disease. By contrast, patients with HLH with infectious or malignant triggers had the poorest survival. Warranting further investigation, most identified DIAP gene variants are currently of unknown significance.
The findings by Chinn et al strengthen the notion that HLH could be the end result of a variety of pathophysiological mechanisms (see figure), carrying both diagnostic and therapeutic implications. The authors make a case for revising the current diagnostic algorithm for children with HLH. As they point out, cellular assays of perforin expression and exocytosis should still constitute first-line investigations in children with suspected HLH. However, their study highlights the clinical need for genetic analysis beyond the scope of lymphocyte cytotoxicity–related genes, particularly in patients lacking mutations in fHLH genes. Crucially, extended genetic analyses can provide additional molecular diagnoses and reveal new mechanisms of predisposition to HLH, increasing pathophysiological understanding and potentially unraveling new therapeutic avenues. Ultimately, genomics can help stratify secondary HLH into molecularly defined phenotypes, creating the opportunity for better-targeted treatments. As a recent example, individuals with activating NLRC4 mutations develop a form of HLH that is characterized by high levels of interleukin-18 (IL-18) and (partially) responsive to IL-1 inhibition.9 Similarly, biological therapy might represent an efficacious approach in targeting additional inflammasome components associated with HLH, as indicated by Chinn et al.
Even after blurring the division between primary and secondary HLH, questions remain regarding the genetic predisposition to HLH. Despite exome sequencing, Chinn et al could not identify a likely genetic explanation in approximately half of the children tested. Additional genetic mutations might have gone undetected. In a cohort of infants with fHLH, >50% of cases were explained by noncoding mutations.10 Facilitated by the fast-falling cost of sequencing, the future of HLH diagnostics will likely see a rapid implementation of whole-genome sequencing. Because of the opportunity to detect noncoding and structural variants, whole-genome sequencing promises to further close the existing knowledge gaps concerning the pathophysiological mechanisms and potential treatments of HLH.
Conflict-of-interest disclosure: The authors declare no competing financial interests.