Abstract
Epstein Barr virus (EBV) has been implicated in the lymphomagenesis, particularly of HIV-related lymphomas. EBV-infected cells are able to share miRNAs with other cells via exosomes and certain EBV-miRNAs were found in cells not infected with the virus but from patients with increased levels of EBV copies in plasma such as HIV-infected individuals. Thus, EBV-miRNAs might be detected in the plasma and might be relevant for the diagnosis and follow-up of HIV-related lymphomas.
This case-control study included a group of 26 HIV-infected patients with lymphoma that responded to treatment and a control-group of HIV-infected subjetcs without lymphoma matched for age, gender, CD4-lymphocyte counts, plasma HIV viremia and time from HIV infection. We evaluated the presence of 43 EBV-miRNAs in frozen plasma samples from patients with lymphoma at diagnosis (Dx-group, N=26), one year before diagnosis (Pre-group, N=15) and complete response (CR-group, N=26). RNA was extracted from 200µl of plasma using Trizol LS (Ambion, Carlsbad, CA) and cel-miR-39 was used as spike-in. The detection of EBV-miRNAs was assessed by QRT-PCR in a BioMark™ HD System of Fluidigm (San Francisco, CA) using TaqMan® microRNA assays and TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA). The following clinical and biological parameters were collected from records: age, gender, ECOG score, extranodal and bulky disease, B symptoms, Ann-Arbor stage, serum lactate dehydrogenase (LDH) and beta2-microglobulin, International Prognostic Index (IPI), HCV and HBV serology, history of opportunistic infection and of AIDS-defining illnesses, onset of combination antiretroviral therapy, CD4-lymphocyte counts, plasma HIV viremia. McNemar's and Wilcoxon tests were used for paired comparisons between groups. P-values of less than .05 were considered statistically significant.
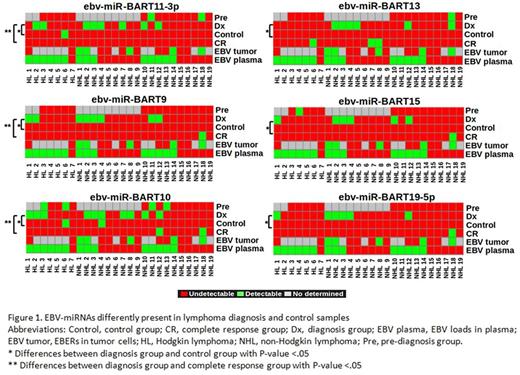
Overall, 30 EBV-miRNAs were detected in the Dx-group whereas only 8 EBV-miRNAs where found in the control-group and 19 in the CR-group. EBV-miRNAs were detected more frequently in the Dx-group (17 out of 26) than in the control-group (7 out of 26) (P =.002) and in the CR-group (11 out of 26) (P=.02). The presence of 6 EBV-miRNAs (ebv-miR-BART11-3p, ebv-miR-BART9, ebv-miR-BART10, ebv-miR-BART15, ebv-miR-BART31, and ebv-miR-BART19) could be associated with the lymphoma condition, since they were detected significantly different in the Dx-group and the control-group (Figure 1). Importantly, 3 of those EBV-miRNAs (ebv-miRBART11-3p, ebv-miR-BART9 and ebv-miRBART10) were found to be significantly less frequent in the CR group, reinforcing the idea that they could be derived from lymphoma cells and be used as biomarkers for diagnosis and follow up of HIV-related lymphoma. EBV-miRNAs cannot be used as early biomarkers of lymphoma in HIV-infected patients because none of the 3 EBV-miRNAs associated with lymphoma was significantly found in samples before diagnosis. The detection in plasma of ebv-miR-BART11-3p, ebv-miR-BART10 or ebv-miR-BART9 was not associated with having tumor cells infected with EBV (Figure 1). Only the detection of ebv-miR-BART9 was associated with detectable EBV loads [7/14 (50%) vs. 1/12 (8.3%), P=.036] (Figure 1). The following were the statistically significant associations between the presence of the 3 potential EBV-miRNA lymphoma biomarkers and the clinical-biological features studied. The presence of ebv-miR-BART10 in plasma was associated with IPI>2 [8/14 (57.1%) vs. 1/9 (11.1%), P=.004] and ECOG>2 [8/14 (57.1%) vs. 1/11 (9.1%), P=.011]. Also, more patients with ebv-miR-BART11-3p expression in plasma had ECOG>2 than patients without it [7/13 (53.8%) vs. 1/12 (8.3%), P=.030].
Ebv-miR-BART11-3p, ebv-miR-BART9 and ebv-miR-BART10 can be detected in the plasma of patients with HIV-related lymphoma, independently of the presence of EBV in tumor cells or of EBV plasma viremia. They are potential biomarkers of lymphoma to be used in the diagnosis and follow up of HIV-infected subjects with the advantage of being easily determined by QRT-PCR employing noninvasive procedures.
This work was supported in part by 2014 SGR225 (GRE) from CERCA Programme/Generalitat de Catalunya, and by funds from Josep Carreras International Foundation, "la Caixa" Foundation, DKMS, and Celgene Spain.
Baptista: DKMS: Research Funding; la Caixa Foundation: Research Funding; Carreras International Foundation: Research Funding; CERCA Programme/Generalitat de Catalunya: Research Funding; Celgene Spain: Research Funding. Sancho: Roche: Honoraria; Laboratorios Servier: Consultancy; Mundipharma: Honoraria; Kern Pharma: Honoraria; Gilead: Honoraria; Celgene: Honoraria; Janssen: Honoraria. Ribera: Celgene: Honoraria; Janssen: Honoraria; Gilead: Honoraria; Amgen Inc.: Research Funding, Speakers Bureau; Pfizer: Research Funding, Speakers Bureau; Incyte: Research Funding, Speakers Bureau; ARIAD: Research Funding, Speakers Bureau; Roche: Honoraria. Navarro: Celgene Spain: Research Funding; DKMS: Research Funding; la Caixa Foundation: Research Funding; Josep Carreras International Foundation: Research Funding; CERCA Programme/Generalitat de Catalunya: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.