In this issue of Blood, the groups of Louissaint et al1 and Schmidt et al2 report on the genetic landscape of pediatric-type follicular lymphoma (PTFL). Their studies confirm suspicions that PTFL represents a biologically distinct form of lymphoma, with fewer recurrent genetic alterations than typical FL. Because this group of patients has an excellent prognosis, accurate and reliable identification of PTFL is important to minimize the prospect of unnecessary treatment.
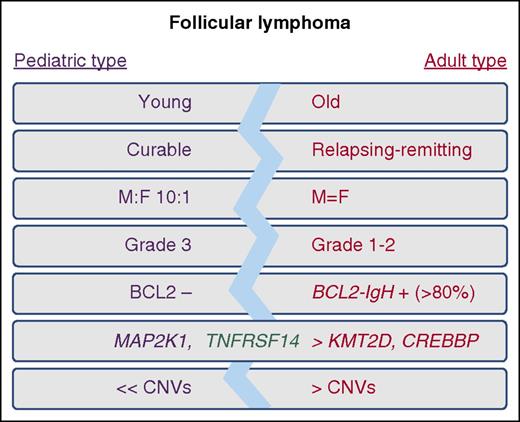
Features of pediatric-type and typical adult-type follicular lymphoma. CNV, copy number variation; F, female; M, male.
Features of pediatric-type and typical adult-type follicular lymphoma. CNV, copy number variation; F, female; M, male.
The 2016 revision of the World Health Organization classification of lymphoid neoplasms promoted pediatric follicular lymphoma (FL) from a provisional to a definitive entity, renaming it PTFL to reflect the incidence in both children and adults.3 Although sharing some features with typical adult FL, PTFL is both histologically and clinically distinct, with blastoid cytology, a clear male predominance (10:1), the majority of patients presenting with localized stage 1 lymphadenopathy, a high proliferation index, and exceedingly good response rate either to local excision or minimal chemotherapy4 (see figure). Going forward, and given the existence of PTFL in adults, it is important to distinguish these different endotypes, irrespective of age, to appropriately stratify patients and deescalate therapy where possible. Although it has been recognized that IgH-BCL2/t(14;18) and overexpression of BCL2 protein is absent in PTFL,4 these 2 new studies offer important insights into the genetic makeup of PTFL and provide compelling evidence to support the notion that PTFL is a distinct entity with unique molecular features.
Both Louissaint et al and Schmidt et al used standard next-generation sequencing and copy number profiling approaches to generate a mutational landscape of PTFL in a combined series of 68 patients. The data are remarkably consistent across studies, with PTFLs deemed genetically light, accumulating fewer recurrent mutations or copy number alterations than typical FL. Schmidt et al rule out the possibility of a close genetic relationship linking PTFL and BCL2-negative typical FLs by providing a direct comparison of mutations in these lymphomas. Both studies confirm the previous report by Martin-Guerrero et al5 of a high frequency (>25%) of TNFRSF14 mutations in PTFL and their common association with deletion or copy number–neutral loss of heterozygosity of chromosome 1p, lesions that also characterize typical FL. However, this is where the genetic semblance diverges, with PTFL having a reduced incidence of mutations in the histone methyltransferases KMT2D and EZH2 and the acetyltransferase CREBBP. Although each of these lesions individually affect B-cell biology, their absence in PTFL is noteworthy because it may suggest a link between the presence of epigenetic mutations and the relapsing clinical course of typical FL.
Moreover, phylogenetic analysis6-8 demonstrated that mutations in the key epigenetic regulators represent early events in FL, and their absence along with the lack of t(14;18) suggests that the initiating events of PTFL are wholly distinct. Critically, the whole exome sequencing data in 22 PTFLs by Louissaint et al identify recurrent mutations in different components of the MAPK pathway (MAP2K1 [43%], MAPK1 [9%], and RRAS [4.5%]), implicating activation of the MEK/ERK pathway in lymphomagenesis. Although these mutations are not unique to PTFL and have been reported in rare cases of B-cell lymphomas, including hairy cell leukemia-variant,9 they provide, as the authors rightly propose, an objective parameter to support the diagnosis of PTFL.
Altogether, we must not be complacent in diagnosing PTFL and the possibility of it masquerading as typical FL. This diagnostic challenge was highlighted perfectly by 2 PTFL cases in the Louissaint et al study that presented with mutation profiles analogous to typical FL, suggesting that a reliance on clinical diagnostic criteria alone may not be sufficient in all cases, particularly within the adult population. Further collaborative studies are now needed to better define the frequency of these cases and consider if PTFL patients need to be accounted for in ongoing efforts at developing a clinico-genetic risk score in FL.10 Indeed, although the genomic landscape of typical FL may be better appreciated, these new studies are excellent examples of the significant insights gained from studying rarer subsets of FL.
Conflict-of-interest disclosure: The authors declare no competing financial interests.