Abstract
Background: Chronic lymphocytic leukemia (CLL) is associated with significant genomic heterogeneity that might explain its variable clinical course. Although there are established prognostic genomic features already detected by fluorescence in-situ hybridization (FISH), next generation sequencing (NGS) technologies have uncovered a myriad of novel mutations in CLL that may influence both prognosis and treatment decisions (Landau DA et al. 2015). Recently commercial NGS panels are actively used outside of the research spectrum. This study describes the mutational landscape of unselected consecutive CLL patients analyzed using commercial NGS platforms at a single academic institution as a part of routine clinical care.
Materials and methods: Using Total Cancer Care (TCC) and Moffitt Cancer Center (MCC) CLL databases (N=1,267 patients) we retrospectively identified a subset of 148 subjects from January 2014 to May 2016 with confirmed CLL diagnosis and an available commercial NGS panel (Genoptix® and FoundationOne®). Molecular testing was done on bone marrow biopsies and/or peripheral blood samples either during the first clinic visit, or at time of disease progression. We used descriptive statistics to annotate demographic characteristics, disease prognostic features, mutation frequencies, and the class and type of molecular changes found in the cohort. We utilized standard statistical methods (Chi-square) to assess associations between known prognostic factors (age, CLL-IPI score, IGHV mutational status, B2M, ZAP-70 and CD38+ status, and FISH detected mutations) and identified mutations. Time to first treatment was estimated with the Kaplan-Meier method and curves compared with log-rank test. All statistical analyses where done using SPSS v24.0
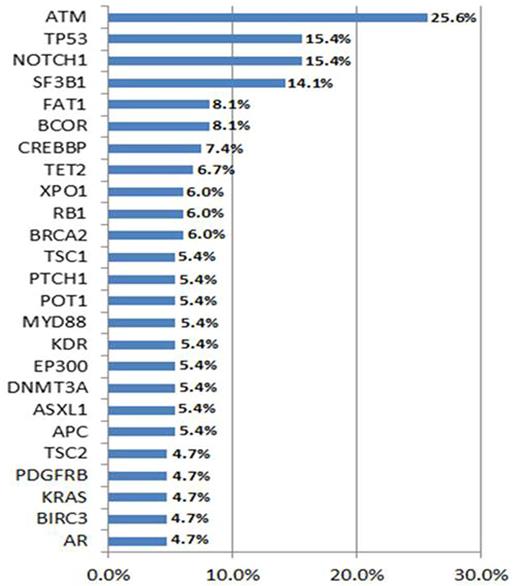
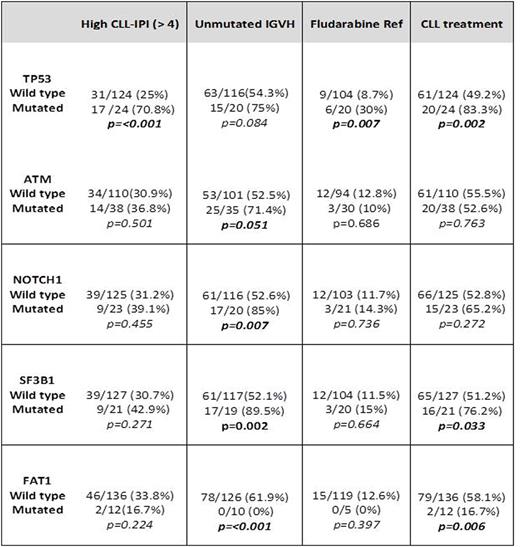
Results: From our cohort of 148 CLL patients with an available commercial NGS panel, 110 and 38 were sequenced using Genoptix® and FoundationOne® panels, respectively. At the time of molecular sequencing 48 (32.3%) had high or very high CLL-IPI scores, 77 (52%) had unmutated IGHV, 22 (14.8%) harbored del11q, 19 (12.8%) had del17q and 10 (6.7%) had a complex karyotype. Seventy (47.2%) patients were treatment naïve and 59 (40%) had received CLL directed therapy before NGS testing. Of the previously treated patients, 56% received fludarabine-based regimens and 27% received at least one B-cell receptor inhibitor. The mean number of mutations in our patient cohort was 6.3 (3.1 per Genoptix® and 9.5 per FoundationOne®). Most common mutations types were missense (74.7%) and frameshift (14%). Most frequently mutated genes (≥7 cases) were: ATM (25.6%), TP53 (15.5%), NOTCH1 (15.5%), SF3B1 (14.1%), FAT1 (8.1%), BCOR (8.1%) and CREBBP (7.4%) (Figure 1). TP53 mutations had higher CLL-IPI scores (p<0.001), del17q (p<0.001), previous fludarabine based therapy (p<0.009), fludarabine resistance (p=0.016) and shorter TFT in comparison to TP53wt (16.5 vs. 64.2 months, p=0.002). Interestingly, FAT1 mutation was universally correlated with mutated IGHV status (p<0.001), and lower probability CLL-directed treatments (p=0.006). Table 1 shows correlation of general characteristics and other mutated genes.
Conclusions: NGS, through validated commercial gene platforms, is readily available outside of clinical trials for CLL patients for genomic risk assessment and could accurately replicate findings of earlier reports. Challenges remains in regards of cost, adequate bioinformatics support and accessibility that restrict its availability to academic centers.
Percentage of patients with the most common mutated genes
Percentage of patients with the most common mutated genes
Nodzon:Novartis: Speakers Bureau. Chavez:Janssen: Speakers Bureau. Pinilla-Ibarz:Abbvie: Consultancy, Speakers Bureau; Pharmacyclics: Consultancy, Speakers Bureau; Gilead: Consultancy, Speakers Bureau; Novartis: Consultancy; Novartis: Consultancy; Janssen: Consultancy, Honoraria; Gilead: Consultancy, Speakers Bureau; Abbvie: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.