Abstract
Background. Diffuse large B-cell lymphoma (DLBCL) is one of the most aggressive types of B-cell lymphoma with high heterogeneity, accounting for 30-40% of newly diagnosed non-Hodgkin lymphoma (NHL), and dysfunction of epigenetic regulation has been found as a common and important feature of B cell lymphomas. To identify epigenetic associated genes mutations in DLBCL, including KMT2D, CREBBP, EP300, EZH2 and MEF2B, we sequenced tumour DNA from 226 Chinese DLBCL cases by applying next generation sequencing technology (NGS). A total of 679 consecutive Chinese patients with previously untreated DLBCL at our institution from December 2006 and January 2016 were enrolled in this study, and we assessed the predictive value of clinical and mutational pattern of epigenetic associated genes in a large single-institution cohort of these patients.
Methods. Genomic DNA was extracted from 226 subjects with DLBCL formalin-fixed paraffin-embedded tumor tissue, using a QIAamp DNA FFPE Tissue Kit (Qiagen). Specific primers, producing amplicons about 200 bp at the coding regions of the genes of interest , were designed at the UCSC website (http://genome.ucsc.edu/cgi-bin/hgGateway ). Microfluidic PCR reactions ran in a 48 ¡Á 48 Access array system (Fluidigm) with FastStart High Fidelity PCR system (Roche) and high-throughput DNA sequencing was performed on Illumina Genome Analyzer IIx (GAIIx) and HiSeq2000 systems, according to the manufacturer's instructions. SAMtools version 0.1.19 was used to generate chromosomal coordinate-sorted bam files and to remove PCR duplications. Sequences for epigenetic associated genes were obtained from the UCSC Human Genome database, using the corresponding mRNA accession number as a reference, and those containing splice-site, nonsense or coding-region indel mutations, were selected for Gene Ontology analysis. All of the results were also confirmed by Sanger sequencing. Baseline characteristics of patients were analysed using two-sided c2 test. Overall survival (OS) was estimated using the Kaplan-Meier method and compared by log-rank test. Univariate hazard estimates were generated with unadjusted Cox proportional hazards models. Covariates demonstrating significance with P<0.100 on univariate analysis were included in the multivariate model. Statistically significance was defined as P<0.05. All statistical analyses were carried out using Statistical Package for the Social Sciences (SPSS) 20.0 software (SPSS Inc., Chicago, IL, USA).
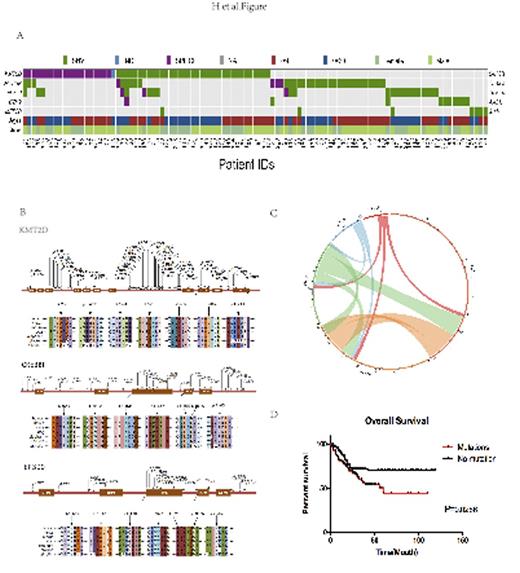
Results. Overall, 105 of 226 Chinese DLBCL cases were identified to have at least one mutation in epigenetic regulator genes. Somatic mutations in KMT2D were most frequently observed (24.3%), followed by CREBBP, EP300, EZH2 and MEF2B (15.5%,10.6%,4.4% and 2.2%, respectively)(Figure1,A,B). Association of mutated genes according to the conceptual classification. Circos plot of mutated genes according to the function is shown, and overlap mutations between epigenetic regulator genes mutations were frequently observed (Figure1, C). Clinically, mutation-positive DLBCL patients presented shorter OS than patients those without mutations (P=0.0286, Figure 1,D) among 226 DLBCL cases. A total of 679 Chinese DLBCL cases were enrolled in univariate analysis, and R-IPI, Complete Remission (CR), epigenetic related mutations were significant prognostic factors for OS. In further multivariate analysis, R-IPI (RR=2.72,95%CI=1.619-4.567,P<0.000), CR (RR=0.129,95%CI=0.076-0.219,P<0.000), epigenetic related mutations (RR=1.605,95%CI=1.007-2.557,P=0.046) are independent prognostic factor for OS.
Conclusion. Our study provided the mutational spectrum of epigenetic regulator genes in DLBCL, and the relationships between mutations and clinic suggested some therapeutic efficiency.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.