In this issue of Blood, Deininger et al demonstrate that BCR-ABL1 mutations including compound mutations are not a major cause of resistance to ponatinib for chronic myeloid leukemia (CML) patients resistant to multiple tyrosine kinase inhibitors (TKIs).1
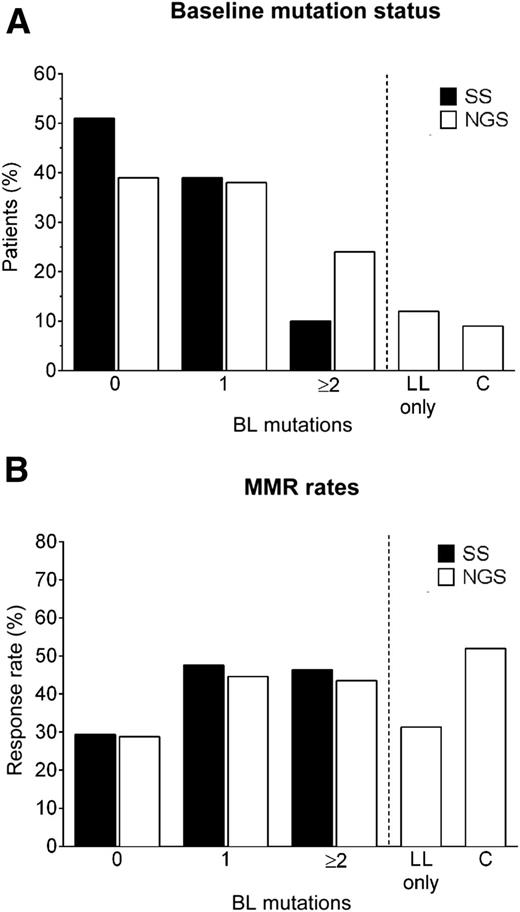
BCR-ABL1 mutation assay by SS and NGS at baseline (A). Rates of major molecular response (MMR) rates according to baseline mutation status (B). BL, baseline; C, compound; LL, low-level. See Figure 2A,C in the article by Deininger et al which begins on 703.
BCR-ABL1 mutation assay by SS and NGS at baseline (A). Rates of major molecular response (MMR) rates according to baseline mutation status (B). BL, baseline; C, compound; LL, low-level. See Figure 2A,C in the article by Deininger et al which begins on 703.
TKIs of the BCR-ABL1 oncogenic protein have revolutionized the therapy of CML and undoubtedly represent the gold standard for the treatment of this disease. There are now 5 different TKIs approved for first- and/or second-line CML therapy, giving multiple options for starting and switching therapy for nonoptimal responders. However, there are still CML patients who become sequentially resistant to different TKIs. BCR-ABL1 mutations able to circumvent the inhibition of the BCR-ABL1 tyrosine kinase activity have been recognized as a major cause of resistance to TKIs, but the spectrum of the resistant mutations varies between different TKIs.2 Ponatinib, a third-generation TKI, is so far the only registered TKI that shows activity against all clones expressing mutant BCR-ABL1 proteins at a single residue, including those derived from the T315I-mutated allele, which is able to confer resistance to all other approved TKIs.3 In this issue of Blood, Deininger et al report new interesting observations about the role of mutations in determining resistance to ponatinib and also provide new information about the biology of the Ph-positive leukemias.1
They used 2 different methods to assess the presence of BCR-ABL1 mutations, the traditional Sanger sequencing (SS) and next-generation sequencing (NGS). SS is characterized by a low sensitivity and able to detect a mutated clone only if this represents at least 20% of the resistant cells. NGS is a much more sensitive method, also able to detect the so-called compound mutations (ie, variants containing ≥2 mutations on the same BCR-ABL1 allele). They studied at baseline 267 chronic-phase CML patients enrolled in the PACE trial, a phase 2 study designed to assess the efficacy of ponatinib in a series of patients resistant or intolerant to imatinib and to nilotinib and/or dasatinib or with the T315I mutation.4 Indeed in this trial >90% of the CML cases were resistant to ≥2 TKIs and 60% to ≥3 TKIs. As expected, they found that compared with SS, the NGS sensitive sequencing technique was able to reveal a higher incidence of cases with single mutations as well as cases with multiple mutations and compound mutations. At baseline, SS detected 161 mutations in 131 patients, whereas NGS detected 266 mutations in 163 patients, including the 161 mutations detected by SS and additional 73 mutations present only at low level (<20%, low-level mutations).1 Furthermore, NGS was able to find multiple mutations in 23% of the cases compared with only 10% detected by SS (see figure, part A), and a high percentage of these cases were apparently carrying compound mutations.1 Because it was previously shown that many compound mutations could be artifacts caused by polymerase chain reaction–mediated recombination,5 Deininger at al developed and applied an algorithm that predicts the frequency of false-positive compounds mutations. They found that 88% of possible compound mutations were most likely false positives and that only 9% of patients (42% of patients with multiple mutations) had true compound mutations at baseline (see figure, part A).1 True compound mutations involved one mutation classically implicated in TKI resistance, underlining that the presence of particular mutations makes the resistant clones develop an additional mutation on the same allele, and this may occur more frequently in patients treated sequentially with multiple TKIs such as in the PACE trial.
In vitro experiments had previously shown that compound mutations involving the T315I variant were able to confer resistance to ponatinib.6 Moreover, in a cohort of 64 Ph-positive acute lymphocytic leukemia or CML patients treated in the PACE trial or in the ponatinib expanded access program, compound mutations were reported to occur quite frequently during ponatinib treatment, particularly in more advanced phases of the disease.6 Quite the opposite, the results of Deininger et al show that the presence of true compound mutations in pluri-resistant CML patients in chronic phase is quite low and that they do not represent a major cause of primary resistance to ponatinib. Irrespective of the method used (SS or NGS) to assess the presence of mutations, the responses observed in chronic-phase CML patients enrolled in the PACE trial and their durability could not be predicted on the basis of the mutational status (presence and type of mutation) determined at baseline (see figure, part B).1 Furthermore, reanalyzing with the SS method postbaseline samples of 129 patients who discontinued ponatinib treatment of poor response or for adverse events, the expansion of clones harboring new mutations not detected at baseline by NGS, including compound mutations, were found in only 8 patients, suggesting again that mutations, including compound mutations, are not major drivers in determining resistance to ponatinib.
These findings reveal new aspects of the role that mutations and genomic instability have in determining progressive resistance to TKI therapy. During the initial phase of the disease, resistance to TKI therapy seems to be mainly dependent on the selection and expansion of clones harboring BCR-ABL1 mutations that allow the persistence of the BCR-ABL1 tyrosine kinase activity. At later stages of the disease, resistance to TKI therapy seems to rely mainly on the activation of BCR-ABL1–independent mechanisms,7 which involve oncogenic pathways other than BCR-ABL1 that make BCR-ABL1 TK activity redundant and dispensable. According to this interpretation, the efficacy of ponatinib in the patients resistant to multiple TKIs could be explained not only by its ability to inactivate BCR-ABL1 mutations, but also by its aptitude to suppress other oncogenic routes than those strictly dependent on the BCR-ABL1 TK activity. Indeed ponatinib is known to be a multikinase inhibitor.8 This consideration may support a more extensive investigation of the efficacy of ponatinib, either in early chronic-phase CML, because of its ability to suppress mutations and controvert genomic instability, as well as in Ph-negative hematologic malignancies, owing to its multitarget activity.9,10
Conflict-of-interest disclosure: The authors declare no competing financial interests.