In this issue of Blood, Zhang et al show that genes of pluripotent stem cells coding for platelet surface glycoproteins may be changed to encode platelet-specific alloantigens, and that such antigens are efficiently expressed on the surface of the manipulated cells.1 The significance of this work is that such cells can be useful tools for the diagnostic studies of antibody-mediated platelet destruction.
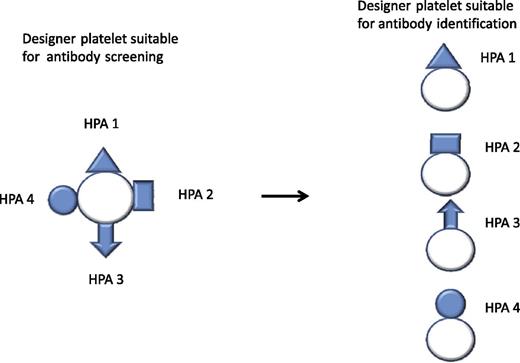
Designer platelet expressing a variety of clinically important antigens for the initial antibody investigation. Further differentiation of the antibody specificities by designer platelets expressing only 1 or a limited array of antigens.
Designer platelet expressing a variety of clinically important antigens for the initial antibody investigation. Further differentiation of the antibody specificities by designer platelets expressing only 1 or a limited array of antigens.
Several platelet membrane glycoproteins exist as biallelic isoforms, differing by only a single amino acid, and some of these are immunogenic. Alloantibodies to platelet-specific antigens are responsible for the most severe cases of neonatal thrombocytopenia, posttransfusion purpura, and platelet transfusion refractoriness.2 The clinical consequences of those conditions may be severe bleedings, with intracranial hemorrhage, lifelong neurologic damage, or prenatal death as the most feared outcomes. To detect and identify those conditions, accurate platelet alloantigen typing and determination of antibody specificity are of considerable importance. Once the diagnosis is established, the provision of compatible platelets for transfusion becomes possible.
Platelet alloantigen phenotyping is mainly limited to human platelet alloantigen (HPA)–1a typing, as prototype antisera for the other allotypes are rare and difficult to obtain. By introduction of easy-to-use genotyping techniques,3 the problem with platelet antigen typing is overcome. Because of genetic polymorphism or point mutations, discrepancy between pheno- and genotyping can occur and give a false interpretation of the typing results; however, such cases are rare.4
Determination and identification of platelet-specific antibodies still remains a challenge and is hampered by technical complexities. Labor-intensive and time-consuming immunoassays are the main tools for antibody identification but represent the gold standard in diagnostic activity.5 Freshly drawn human platelets are still used as central components for diagnoses, although platelets from several donors are necessary to create a representative repertoire of clinically relevant antigens. Some antigens are rare and not readily available, and some are unstable upon storage. Another complicating aspect is that some individuals have a mixture of several platelet-reactive antibodies.
Zhang et al address these challenges to accurately determine platelet-specific antibodies. They have used a novel technology of gene editing to create human platelet progenitor cells expressing platelet-specific antigens of low frequency.
To test this, a proof-of-concept study was conducted to create HPA-1a– and HPA-1b–specific platelets suitable for flow cytometric analysis. The expressed protein entities are recognized by the corresponding human antibodies. By using this technique, creation of platelet progenitors or platelets expressing other clinically significant antigens is currently in progress. To avoid reactivity of unwanted antibodies with the designer platelets, the encoding genes can be manipulated; so, for instance, HLA antigen expression may be blocked by introduction of a stop codon in the β2 microglobulin gene. For diagnostic purposes, designer platelets expressing a variety of clinically important antigens could be designed for the initial study. In case antibodies are identified, further differentiation of the specificities could be achieved by using designer platelets expressing only 1 or a limited array of antigens (see figure).
The authors also envisage that “antigen-negative” designer platelets may be manufactured in quantities suitable for transfusion of patients with refractoriness because of platelet-reactive antibodies. It remains to be seen if this is feasible. We have to remember that one of the most immunogenic surface molecules of platelets, namely, glycoprotein IIIa, is of crucial significance to the function of platelets.
The work by Zhang et al is a welcome and promising achievement to create a definitive analytic system for platelet antibody identification, which when aligned with the genotyping techniques for platelet antigens, provides a complete set of diagnostic tools.
Conflict-of-interest disclosure: The author is a stock owner of Prophylix Pharma AS, a small biopharma company working to manufacture a prophylactic drug against fetal/neonatal alloimmune thrombocytopenia.