Abstract
Purpose
According to our experience, ethnic difference in acute myeloid leukemia (AML) is considerable, especially between Caucasians and Asians. In this study, first we tried to identify genomic signature of Korean AML patients and investigat how these genomic signatures would correlate with clinical parameters. Additionally, we tried to compare genomic signatures of Korean AML patients with that of TCGA.
Methods
We identified specific single nucleotide variant (SNV)'s, small indels and copy number variation (CNV)'s using whole exome sequencing (WES) of acute myeloid leukemia (AML) samples from 103 patients. For detection of SNV's and small indels, we used in-house algorithm Adiscan (https//www.syntekabio.com). For detection of CNV's, we used CONTRA. Clinical parameters considered includes gender, age, bone marrow blast percentage, WBC count at diagnosis, French-American-British (FAB) classification, WHO classification, disease free survival (DFS), reception of allogeneic stem cell transplantation (ASCT) and overall survival (OS). We used TCGA data available on cBio portal.
Results
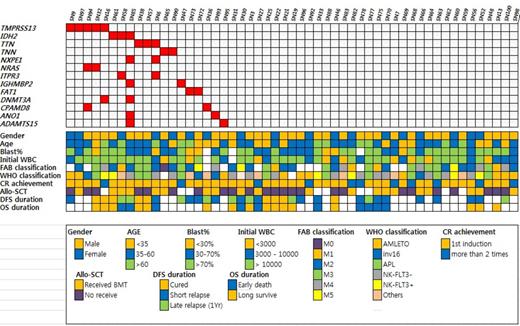
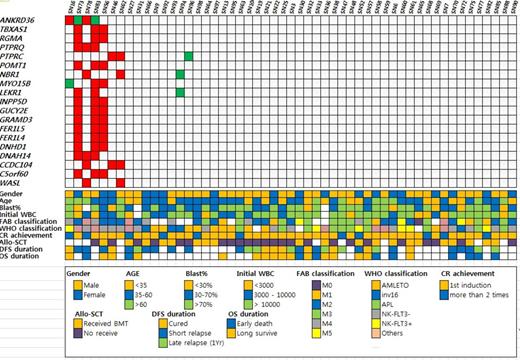
In the analysis, when clustering analysis was performed, gender did not have correlation with genetic signatures both in terms of SNV's, indels and CNV. For patient's age, genetic signature of middle-aged patients (age between 35-60) was rather diverse compared to the young patients (age <35) and old patients (age>60). From phenotypical perspective, M4 and M5 AML by FAB had common genetic signatures, which was in contrast to M2 and M3 disease. It is of note that M3 disease does have heterogeneous genetic signature. When the prognosis of AML patients were considered, patients with excellent prognosis (complete remission for more than 3 years) had rather homogenous genetic clustering, while patients with poor prognosis (relapse in 1 year) had very heterogeneous genetic clustering, which implicate complexity of disease relapse mechanism. Commonly mutated non-synonymous genes include TPMRSS13, IDH2, TTN, TNN, NXPE1, NRAS, FAT1, DNMT3A, ANO1, and ADAMTS15. Copy number changes were observed in genes including PTPRC, PTPRQ, NBR1, and WASL. TPMRSS mutation seemed to have prognostic value but further validation is necessary. Compared to TCGA, TMPRSS13 mutation and TNN mutation were unique SNVs found in Korean AML. On the other hand, frequency of IDH2, NRAS, and DNMT3A was considerably low in Korean AML patients.
Conclusion
Korean AML patients have their own genetic signature which is distinct to that of Caucasians, which supports ethnic difference of AML. TMPRSS13 and TNN mutation in Korean AML patients are noticeable.
Commonly found SNV's found by WES in Korean AML patients and its relation with clinical parameters
Commonly found SNV's found by WES in Korean AML patients and its relation with clinical parameters
Commonly found CNV's found by WES in Korean AML patients and its relation with clinical parameters
Commonly found CNV's found by WES in Korean AML patients and its relation with clinical parameters
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.