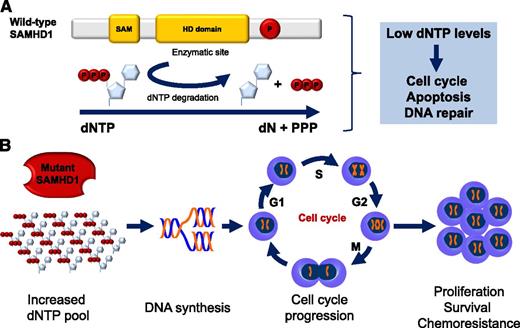
Wild-type SAMHD1 functions and predicted functional consequence of SAMHD1 mutations in CLL. (A) Structure and cellular functions of wild-type SAMHD1. Representative schematic of the important domains of SAMHD1. The enzymatic activity of SAMHD1 is promoted by the HD domain. SAMHD1 degrades dNTPs into deoxynucleosides (dN) and inorganic triphosphates (PPP). Low levels of dNTPs decrease DNA synthesis, slow down cell cycle progression, promote apoptosis, and favor DNA repair. SAM, sterile α motif; P, putative phosphorylation site. (B) Predicted functional consequence of SAMHD1 mutations in CLL. SAMHD1 loss of function is predicted to result in increased dNTP cellular levels, DNA synthesis, cell cycle progression, and ultimately, proliferation, survival, and chemoresistance.
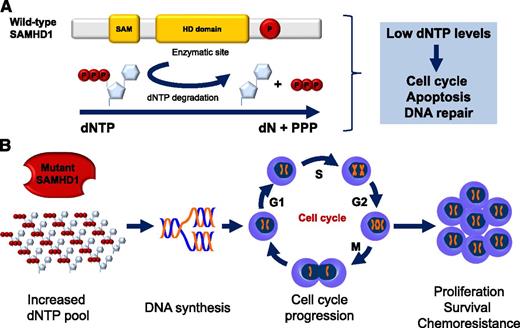
Wild-type SAMHD1 functions and predicted functional consequence of SAMHD1 mutations in CLL. (A) Structure and cellular functions of wild-type SAMHD1. Representative schematic of the important domains of SAMHD1. The enzymatic activity of SAMHD1 is promoted by the HD domain. SAMHD1 degrades dNTPs into deoxynucleosides (dN) and inorganic triphosphates (PPP). Low levels of dNTPs decrease DNA synthesis, slow down cell cycle progression, promote apoptosis, and favor DNA repair. SAM, sterile α motif; P, putative phosphorylation site. (B) Predicted functional consequence of SAMHD1 mutations in CLL. SAMHD1 loss of function is predicted to result in increased dNTP cellular levels, DNA synthesis, cell cycle progression, and ultimately, proliferation, survival, and chemoresistance.
SAMHD1 encodes the sterile α motif and histidine-aspartic domain (HD) containing protein 1, a deoxynucleoside triphosphate triphosphohydrolase that degrades the intracellular pool of deoxynucleoside triphosphates (dNTPs) into their deoxynucleosides and inorganic triphosphates (see figure).3,4 The final consequence of SAMHD1 activity is the depletion of the cellular pool of dNTPs, which are required for DNA polymerase functioning and, ultimately, for DNA synthesis (see figure).5
Before being identified as a cancer gene by Clifford et al, SAMHD1 was already known in hereditary genetics because it was constitutively mutated in Aicardi-Goutières syndrome, an inherited autoimmune inflammatory encephalopathy.6 Clifford et al initially pointed to SAMHD1 as a potential CLL gene after their pivotal clinical observation of early-onset CLL in a young adult subject affected by Aicardi-Goutières syndrome. By genome sequencing, CLL cells of this patient were devoid of somatic genetic lesions that are well-known drivers of this leukemia, thus supporting the notion that the germ-line homozygous mutation of SAMHD1 in this patient was sufficient to cause CLL. In parallel, the same authors and others have shown that somatically acquired SAMHD1 mutations also exist in CLL as founder genetic events.7,8
Based on these observations, Clifford et al propose that SAMHD1 mutations could play an oncogenic role in the early pathogenesis of CLL and provide several lines of experimental evidence in support of this hypothesis. First, SAMHD1 is recurrently affected in 3% to 11% of CLL, depending on the case mix of the series investigated, with a prevalence that is in the range of that of well-established driver genes in this leukemia. Second, SAMHD1 mutations are generally represented in the entire CLL clone, indicating that they are early events in CLL clonal evolution. Third, SAMHD1 mutations include disrupting events and often couple with loss or acquired uniparental disomy of the second allele. This molecular pattern recapitulates a typical double hit mechanism of tumor suppressor gene inactivation selected to disrupt SAMHD1. Consistently, SAMHD1 molecular lesions associate with markedly reduced mRNA and protein expression in CLL cells. Fourth, a fraction of SAMHD1 mutations is common to both CLL and Aicardi-Goutières syndrome, where these variants have well-established functional consequences.1,6 Finally, SAMHD1 is involved in the response to DNA damage and in restraining cell survival and proliferation, with the latter cellular process being favored by the mutant form of SAMHD1.
Overall, these results point to a tumor suppressor role of SAMHD1 in CLL. However, several biological questions remain to be elucidated. Do SAMHD1 mutations affect the biochemistry of the protein and its function as deoxynucleoside triphosphate triphosphohydrolase? Which are the specific mechanisms mediating the antiproliferative and proapoptotic effects of SAMHD1 and how are these mechanisms turned off by mutations? What is the role of SAMHD1 in normal B-cell homeostasis? Does SAMHD1 conditional deletion promote in vivo B-cell tumor development?
The study by Clifford et al also opens new questions in the field of CLL predisposition. Much epidemiologic evidence is in support of the genetic susceptibility of CLL, and genome-wide association studies have allowed the identification of several previously unexpected CLL risk loci and genes.9 The discovery by Clifford et al of a SAMHD1 germ-line mutation as a CLL driver points to SAMHD1 and its polymorphisms as attractive candidates to be further explored in the search of risk alleles for CLL.
From a clinical standpoint, Clifford et al propose SAMHD1 mutations as a potential biomarker of chemoresistance in CLL. This is supported by the clinical observation of poorer responses to first line immunochemotherapy among SAMHD1-mutated CLL patients and by the biological observation that loss of SAMHD1 enhances resistance to chemotherapy-induced DNA damage.1 However, larger and well-designed clinical studies and preclinical models are required to validate SAMHD1 mutations as predictors of treatment refractoriness in CLL and to dissect the biology of chemoresistance associated with these mutations. An intriguing mechanism might link SAMHD1 defects to chemorefractoriness. The cytotoxicity of fludarabine, a cornerstone of current CLL treatment, depends on the rate of its incorporation into tumor DNA in place of dNTPs. Because dNTPs compete with fludarabine for incorporation into DNA, their excess in tumor cells confers resistance to these agents.10 SAMHD1 acts as a negative regulator of the intracellular dNTP pool, and therefore, its defects in CLL cells may cause resistance to fludarabine by favoring the accumulation of cellular dNTPs.
As witnessed by the authors, they are at the beginning of a new story in CLL biology.
Conflict-of-interest disclosure: The author declares no competing financial interests.