Abstract
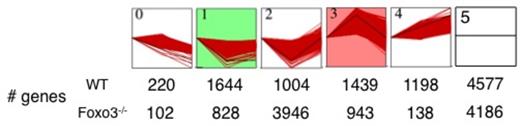
Genes with FPKM>2 from WT and Foxo3-/- samples analyzed with the STEM software, divided into 6 different categories according to their expression profiles during terminal erythroid cell maturation from pro- to polychromatophillic erythroblasts. Genes were then further grouped in 3 subsets: down-regulated, up-regulated and no change. The number of genes in each profile is indicated at the bottom for wild type and Foxo3-/- samples.
Genes with FPKM>2 from WT and Foxo3-/- samples analyzed with the STEM software, divided into 6 different categories according to their expression profiles during terminal erythroid cell maturation from pro- to polychromatophillic erythroblasts. Genes were then further grouped in 3 subsets: down-regulated, up-regulated and no change. The number of genes in each profile is indicated at the bottom for wild type and Foxo3-/- samples.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.