Abstract
It is now clear that traditional response criteria in acute myeloid leukemia based on morphological assessment alone is suboptimal for the prediction of long-term outcome following treatment. While pre-treatment risk stratification based on cytogenetic and molecular correlates of disease biology is highly informative, direct assessment of disease burden after treatment while in traditional complete remission can provide additional prognostic information. Determination of minimal residual disease (MRD) can be performed by a variety of methods; in AML the absence of a unique marker as found in CML and APL has arguably limited the clinical adoption of this higher sensitivity standard. Wilms tumor-1 (WT1) is a commonly over-expressed gene in AML proposed as a standardized MRD assay; unfortunately it is not expressed in 10-20% cases of AML, and is overexpressed to a level necessary for high sensitivity measurements of disease burden in only around 50% of cases.
We performed quantitative reverse transcription polymerase chain reaction (qRT-PCR) to determine the magnitude of overexpression of 65 genes using 30 peripheral blood samples from untreated AML patients and 50 healthy normal donor (ND) controls. From this analysis we selected 11 genes with sufficient overexpression for potential use in AML MRD assessment. We tested the predictive and prognostic capacity of this novel multi-gene array to predict hematological relapse using peripheral blood samples from 18 AML patients either prior to, and/or in the surveillance period following, allogeneic stem cell transplantation (SCT).
De-identified, clinically annotated peripheral blood samples from AML patients who had received allogeneic, non-myeloablative, T-cell depleted SCT from 2004 -2012 collected under IRB-approved protocols at the National Institutes of Health Clinical Center were studied. Expression of 11 genes was assessed using our custom-designed qRT-PCR array, normalized to ABL1 expression and analyzed using the comparative ddCT method After excluding one outlier for each gene, the threshold for positivity was set as the upper limit observed in ND controls. In serial samples, molecular relapse was defined as new gene overexpression seen on two consecutive time-points with the time of relapse defined as first detection of elevation plus 7 days to allow for retesting.
Eighteen patients were studied retrospectively. Average age was 41 years (range 17-72) with 10 female and 8 male. Sixteen patients had available pre-transplant peripheral blood with a minimum clinical follow-up of 10 months and 14 patients had at least 6 months of follow-up samples (including a sample in the three months prior to clinical relapse if applicable).
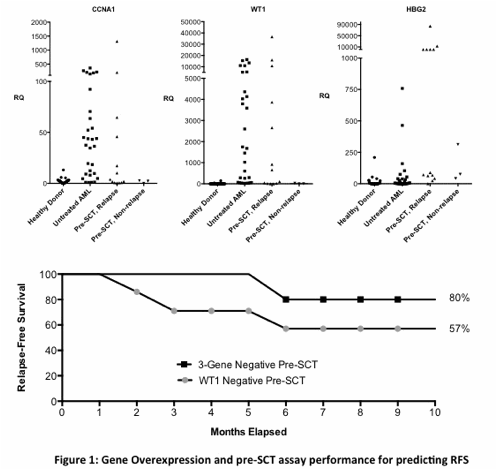
In samples from 16 patients taken prior to SCT, WT1 overexpression was detectable in 9 of the 12 patients who relapsed within ten months after SCT and in none of 4 non-relapsing patients (sensitivity: 75%, specificity: 100%, PPV: 100%, NPV: 57%). In comparison, the use of three genes (WT1, CCNA1 and HBG2) from our multi-gene array correctly predicted 11 of these 12 relapsing patients and was also negative in all non-relapsing patients (sensitivity: 92%, specificity: 100%, PPV: 100%, NPV: 80%). For those nine patients judged to be in clinical remission at the time of SCT, the use of WT1 overexpression alone as a marker of MRD predicted only three of the six relapses in the following ten months (sensitivity: 50%, specificity: 100%, PPV: 100%, NPV: 50%) compared to the identification of five of the six relapses (sensitivity: 83%, specificity: 100%, PPV: 100%, NPV: 75%) using our three gene pre-SCT signature.
In serial surveillance samples from 14 patients following SCT, 7 of the 11 relapsing patients were detected by WT1 overexpression alone compared to 9 detected using our multi-gene signature (sensitivity 64% vs. 82%), with no false positives detected using either test (specificity 100% and PPV: 100% for both methods). The mean time between detection of molecular relapse and clinical diagnosis using WT1 testing was 24 days (range 0-79 days) compared to 46 days (range 0-179 days) using our multi-gene array.
Assessment of minimal residual disease for AML using our multi-gene array is highly sensitive and may improve the ability to predict or detect relapse in both the pre and post SCT settings compared to assays of single gene overexpression. This array will now be tested prospectively.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

