In this issue of Blood, Lechtenberg et al determine the structure of a re-engineered prothrombinase complex built from proteins originally identified in the venom of the Australian eastern brown snake (Pseudonaja textilis).1
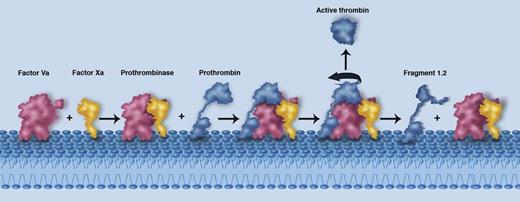
A schematic illustrating the membrane-mediated assembly of the prothrombinase complex (Factor Va in red and Factor Xa in yellow), the proposed interaction with prothrombin (blue), and the subsequent release of active thrombin and fragment 1.2. The individual components are labeled. Professional illustration by Marie Dauenheimer.
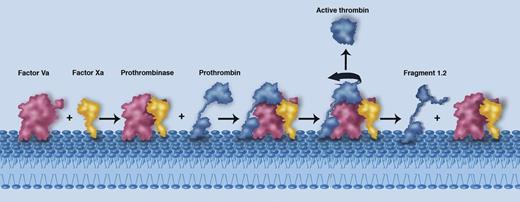
A schematic illustrating the membrane-mediated assembly of the prothrombinase complex (Factor Va in red and Factor Xa in yellow), the proposed interaction with prothrombin (blue), and the subsequent release of active thrombin and fragment 1.2. The individual components are labeled. Professional illustration by Marie Dauenheimer.
A pivotal step in coagulation is the production of thrombin by the prothrombinase complex. The structure of this key enzyme system has, however, long eluded detailed characterization, hampering the development of therapeutic pro- or anticoagulants. The prothrombinase complex represents the engine room of the coagulation system, producing a cloud of active thrombin around the site of tissue injury.2 At a general level, the interaction of factor Va (a large multidomain protein) with the serine protease factor Xa on the membrane surface results in formation of the prothrombinase complex (see figure). The prothrombinase assembly then binds to and cleaves prothrombin at 2 distinct sites (Arg 320 and Arg 271), culminating in the release of soluble active thrombin into the surrounding milieu and promoting the formation of a clot.2
Prothrombinase has presented an exceptionally challenging target for crystallographers, not least because of the lability of the complex and the absolute requirement of phospholipids for complex assembly. Further, electron microscopy experiments have only yielded low-resolution data to date.3 Here Lechtenberg and colleagues neatly surmount these problems and determine the structure of a re-engineered form of pseutarin C, a preassembled soluble snake venom prothrombinase.1 The actual structure determined is between the 2 P textilis components in their preactivated state: factor V in complex with a truncated form of factor X. In an elegant series of experiments, the authors validate the relevance of their system, showing that the relevant forms of pseutarin C that they investigate are capable of cleaving and activating human prothrombin.
The resulting structure provides an exceptional series of insights into the equivalent human system. Broadly, factor X (and by implication Xa) nestles into the interface between 2 key domains of factor V (and again by implication Va): the A2 and A3 domains. An extraordinary feature of the interaction is, however, an extended sequence in the factor V component that reaches out like an embracing arm to encircle the factor X protease domain. Through a series of modeling experiments, the authors then demonstrate the implications of their structure for the human system. Both simple and sophisticated concepts emerge.
At the basic level, the data reveal how factors Va and Xa essentially rise to the same height off the membrane level, thus positioning key surfaces that must interact with one another in the correct locale (see figure). Crucially, the major interactions between the A2 domain of factor V and factor X as seen in the structure are consistent with previous biochemical observations.4 Most notably, these data explain why cleavage and removal of the A2 domain by activated protein C will inevitably lead to complex dissociation and prothrombinase inactivation.5
A key question that remains to be fully understood relates to how prothrombin is captured and cleaved by the prothrombinase complex. Lechtenberg et al, however, are able to interpret their structure to suggest a logical position for the prothrombin component (Figure 3 in Lechtenberg et al1 ). Crucially, the position of the prothrombin protease domain is dictated by the location and bearing of the active site of the factor X moiety. These data suggest that to be placed appropriately, an extended loop sequence (the a1 loop) likely functions to capture the prothrombin target. Electrostatic complementarity further suggests that prothrombin must be orientated such that its Arg 320 site is cleaved first, consistent with biochemical insights.6 Following cleavage at Arg 320, the partially processed intermediate moiety (termed meizothrombin) undergoes the classical rearrangement from the zymogen to the protease (active) form. It is suggested that this conformational change drives repositioning of meizothrombin with respect to the prothrombinase complex such that the Arg 271 site can be cleaved. The latter event releases the active thrombin protease into the surrounding environment (see figure).
The current structure finally provides satisfying insight into how the prothrombinase complex assembles and functions to cleave prothrombin. Exciting additional questions now arise, however. These include addressing how the human system is controlled through factor Va interaction with phospholipids and, furthermore, understanding the precise molecular details of how the prothrombin substrate interacts in 2 distinct positions with the prothrombinase complex.
Snake venoms have long been exploited as a rich source of factors of utility in investigating the coagulation system.7 Here, the weapon of the enemy has once again proved central to understanding human biology. Together, the system developed by the authors of the present study provides a compelling and adaptable platform to determine the complete ternary complex between prothrombinase and prothrombin.
Conflict-of-interest disclosure: The author declares no competing financial interests.