Abstract
Adoptive therapy with T-cell receptor (TCR)–engineered T cells is a promising approach in cancer treatment. While usage of T cells specific for tumor-associated antigens (TAAs) can lead to serious side effects because of autoimmunity, targeting true tumor-specific mutations, such as the products of translocations in leukemias, should reduce such a risk. A potentially ideal target might be the chimeric protein TEL-AML1, which results from the chromosomal translocation 12;21 and represents the most common fusion gene in childhood B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Within the fusion region of TEL-AML1, a single epitope has been described by reverse immunology as immunogenic in HLA-A*0201 restriction settings. As a potential source of TCRs specific for this TEL-AML1 epitope, we have used mice expressing a human TCR-αβ repertoire and human MHC class I. Surprisingly, we have found that, although a specific functional CD8+ T-cell response against this peptide could be evoked, the described epitope was in fact not endogenously processed. Analyses done with a potent antigen-presenting cell line, as well as with purified human proteasomes, support the conclusion that this peptide cannot be proposed as a potential target in immunotherapy of ALL in HLA-A*0201-restricted fashion.
Introduction
Immunotherapy of cancer through adoptive transfer of gene-modified T cells is a promising approach1-3 ; however, the choice of target antigen is decisive for its success.4 Although targeting tumor-associated self-antigens is an approach applicable to a variety of tumors, serious side effects are occasionally seen. Adoptive transfer of autologous peripheral lymphocytes genetically engineered to express TCRs specific for melanocyte differentiation antigens Melan-A/MART-1 and gp100 in melanoma patients led to a partial clinical response, accompanied by autoimmune attack against skin, retina, and inner ear.5 Targeting ERBB2, which is overexpressed in a variety of tumors, through chimeric antigen receptor (CAR)–transduced lymphocytes in a colon cancer patient resulted in a serious adverse event, because of recognition of low levels of ERBB2 on normal lung cells.6 Targeting true tumor specific mutations, such as the products of translocations in leukemias, should reduce such a risk of autoimmunity. The chimeric protein TEL-AML1, resulting from the chromosomal translocation 12;21 represents the most common fusion gene in childhood BCP-ALL,7 which places it among principal target candidates. Furthermore, because the TEL-AML1 fusion protein is an important transforming factor in leukemogenesis,8,9 selection of antigen-loss variants is less likely to occur.
A nonamer peptide derived from the TEL-AML1 fusion region has been described as immunogenic in HLA-A*0201 restriction settings.10 Using reverse immunology approach,11 a peptide which binds to HLA-A*0201 was identified. The peptide was used to induce cytotoxic T-cell (CTL) lines, which recognized autologous leukemic cells and the leukemic cell line REH. Engineering T cells with a TCR, which would specifically recognize this described epitope, should enable us to design a promising adoptive T-cell therapy protocol, called TCR gene therapy.12
As a potential source of TCRs specific for the described TEL-AML1 epitope, we have used a mouse transgenic model for the entire human TCR-α and TCR-β gene loci and human MHC class I, termed ABabDII mice.13 The human TCRs in these mice are HLA-A*0201-restricted, because the mice carry HHD molecule as a transgene, which consists of the HLA-A*0201 molecule with the murine H2-Db α3 domain to enable binding to murine CD8 coreceptor, additionally fused to human β2-microglobulin.14 Previously, we showed that functional specific CD8+ T cells against a variety of described HLA-A*0201-restricted epitopes derived from human tumor antigens could be induced in these mice.13 Therefore, they provide an exquisite tool for testing the immunogenicity of potential human tumor antigen epitopes, as well as for isolation of therapeutic human TCRs.
Surprisingly, we found that the described TEL-AML1 peptide exhibited a very low HLA-A*0201 binding affinity and an anchor-modification had to be introduced to render the peptide immunogenic.15 Moreover, multiple evidence presented here showed that the peptide was not naturally processed, and therefore could not be proposed as a suitable target in immunotherapy of ALL in HLA-A*0201-restricted fashion.
Methods
Mice
ABabDII mice have been described in detail.13 They are transgenic for entire human TCR-α and TCR-β gene loci, as well as for HHD molecule,14 and deficient for the murine Tcr-α and -β chains, as well as for murine β2m and H2-Db genes. The mice used in the study were 2-6 months old and were housed at the Max-Delbrück-Center animal facility. All animal experiments were approved by the Landesamt für Arbeitsschutz, Gesundheitsschutz und technische Sicherheit, Berlin, Germany.
Construction of retroviral vectors for expression of TEL-AML1 minigene, Melan-A/MART-1 and HHD
The nucleotide sequence comprising the TEL-AML1 fusion region (TEL-AML1 minigene): atggtctctgtctccccgcctgaagagcacgccatgcccattgggagaatagcaGaatgcatacttggaatgaatccttctagagacgtc (upper case indicates the fusion nucleotide; italics, the sequence corresponding to the TEL-AML1 peptide), coding for 30 amino acids of the fusion protein, was cloned as an open reading frame (ORF) comprising enhanced green fluorescent protein (EGFP) and a yeast ubiquitin (Ub) moiety into the retroviral vector pMP71.16 This was done in the following way: the TEL-AML1 minigene was first subcloned into the lentiviral vector pRRL.CMV.GFP.sin18,17 which was based on Ub/protein/reference (UPR) technique18 and contained EGFP and the yeast Ub moiety. More precisely, a DNA fragment comprising the sequence of TEL-AML1 and additional 185 bp corresponding to the Ub sequence (placed upstream of the TEL-AML1): cggtaaaaccaatgcattggaagttgaatcttccgataccatcgacaacgttaagtcgaaaattcaagacaaggaaggtatccctccagatcaacaaagattgatctttgccggtaggcagctagaaggcggtagaacgctgtctgattacaacattcagaaggagtccaccttacatcttgtgctaaggctccgcggtggcatggtctctgtctccccgcctgaagagcacgccatgcccattgggagaatagcagaatgcatacttggaatgaatccttctagagacgtctaaaggatccgcg (Geneart) was digested with BstXI and BamHI and subcloned into the same restriction sites of pRRL.CMV.GFP.sin18, so that the resulting segment EGFP-Ub-TEL-AML1 constituted a single ORF. Afterward, the whole ORF (1131 bp) was PCR-amplified (Phusion Hot Start High-Fidelity Polymerase, Finnzymes) with primers designed to introduce NotI and EcoRI restriction sites (forward: atgcggccgccaccatggtgagcaagggcg, reverse: atgaattcttagacgtctctagaaggat; Eurofins MWG Operon), digested with NotI and EcoRI enzymes and cloned into the retroviral vector pMP71.16 The nucleotide sequence of Melan-A/MART-1 (comprising the anchor-modified decamer epitope [italics]): atgccaagagaagatgctcacttcatctatggttaccccaagaaggggcatggccactcttacaccacggctgaagagctcgctgggatcggcatcctgacagtgatcctgggagtcttactgctcatcggctgttggtattgtagaagacgaaatggatacagagccttgatggataaaagtcttcatgttggcactcaatgtgccttaacaagaagatgcccacaagaagggtttgatcatcgggacagcaaagtgtctcttcaagagaaaaactgtgaacctgtggttcccaatgctccacctgcttatgagaaactctctgcagaacagtcaccaccaccttattcaccttaa was cloned into the pMP71 in the similar way: an ORF (1395 bp) consisting of EGFP, Ub and Melan-A/MART-1 was PCR-amplified (Phusion Hot Start High-Fidelity Polymerase, Finnzymes) from the lentiviral vector pRRL.CMV.GFP.sin1819 with primers designed to introduce NotI and EcoRI restriction sites (forward: atgcggccgccaccatggtgagcaagggcg, reverse: atgaattcttaaggtgaataaggtggtggt; Eurofins MWG Operon), digested with the same enzymes and cloned into pMP71.
The HHD gene14 was cloned into pMP71 in the following way: total RNA was isolated from 2 × 107 splenocytes of HHD transgenic mice (RNeasy Mini, QIAGEN) and 2.5 μg were used for cDNA synthesis (SuperScript II, Invitrogen. The HHD gene was amplified by PCR (Phusion Hot Start High-Fidelity Polymerase, Finnzymes) using 125 ng of cDNA and gene-specific primers harboring NotI and BsrGI restriction sites (forward: gcgcggccgccaccatggccgtcatggcgc, reverse: cgtgtacatcacgctttacaatctcggaga, Eurofins MWG Operon). The retroviral vector pMP71-HHD was generated by subsequent ligation of HHD-encoding sequences into pMP71.16
All constructs were verified by sequence analysis (Eurofins MWG Operon).
Cells
NIH/3T3 fibroblasts (CRL-1658; ATCC)) were cultured in DMEM with 10% FBS (PAN Biotech) and 100 IU/mL Penicillin-Streptomycin. NIH/3T3 cells transduced to express HHD, HHD and TEL-AML1 minigene or HHD and Melan-A/MART-1 (designated NIH-HHD, NIH-HHD-TEL-AML1, and NIH-HHD-Melan-A/MART-1) were cultured the same way as NIH/3T3 cells. Plat-E cells were cultured as described.20 T2 cells (CRL-1992; ATCC) were cultured in RPMI with 10% heat-inactivated FBS (PAN Biotech), 10mM HEPES and 1% Penicillin-Streptomycin. Splenocytes and LN cells were cultured in RPMI with 10% heat-inactivated FBS (PAN Biotech), 1mM sodium pyruvate, 100μM MEM nonessential amino-acids, 50μM β-mercaptoethanol and 10 μg/mL gentamicin. All cell culture reagents were purchased from Invitrogen unless otherwise stated.
Generation of NIH-HHD, NIH-HHD-TEL-AML1, and NIH-HHD-Melan-A/MART-1 cell lines
NIH/3T3 cells were retrovirally transduced to express HHD, HHD, and TEL-AML1 minigene or HHD and Melan-A/MART-1. First, ecotropic Plat-E packaging cells were transfected with pMP71-HHD, pMP71-TEL-AML1, or pMP71-Melan-A/MART-1 in the following way: cells were plated at 1 × 106 pro well (in 6-well plates) and the following day 3 μg of DNA was added, dissolved in 500 μL of Opti-MEM medium (Invitrogen) with addition of 10 μL of Lipofectamin 2000 (Invitrogen). Two days later, viral supernatant was collected and filtered (0.45 μm filters). NIH/3T3 fibroblasts were plated at 5 × 104 cells per mL in 24-well plates and spinoculated (32°C, 800g, 90 minutes) on 2 following days with the viral supernatant, 1 mL in total, containing retroviral MP71-HHD, MP71-HHD + MP71-TEL-AML1 or MP71-HHD + MP71-Melan-A/MART-1 particles, with addition of 4 μg/mL polybrene (Sigma-Aldrich). The resulting cells (NIH-HHD, NIH-HHD-TEL-AML1, and NIH-HHD-Melan-A/MART-1) were stained for HHD using anti-HLA A2 antibodies (AbD Serotec) and sorted based on HHD and EGFP expression to > 97% purity on BD FACSAria II.
Antibodies and flow cytometric analysis
The following antibodies were used: CD8a FITC (clone 53-6.7), CD3e APC (clone 145-2C11), purified CD16/CD32 (clone 2.4G2), CD3e Pacific Blue (clone 500A2), CD107a PE (clone 1D4B; BD Biosciences), IFNγ PE (clone AN18.17.24; Miltenyi Biotec), HLA A2 Alexa Flour 647 (clone BB7.2; AbD Serotec), and CD8 APC-eFluor 780 (clone 53-6.7; eBioscience). Multiparametric flow cytometric analysis was performed on BD FACScalibur or FACScanto II. The data were analyzed by FlowJo Version 7.6.2 software (TreeStar).
Peptides and peptide immunization
The peptides used had purity of > 95% and were purchased either from GenScript or JPT. Nomenclature refers to the position of the first amino acid of a peptide in its corresponding protein, as well as to the position of anchor-modification in the epitope. The peptides used for immunizations were: native TEL-AML1 RIAECILGM (TEL-AML1-334, designated TEL-AML1), anchor-modified TEL-AML1 RIAECILGV (TEL-AML1-334-9V, designated TEL-AML1-9V), anchor-modified peptides derived from gp100 YLEPGPVTV (gp100-280-9V, designated gp100) and from Melan-A/MART-1 ELAGIGILTV (Melan-A/MART-1-26-2L, designated Melan-A/MART-1), long native TEL-AML1 MVSVSPPEEHAMPIGRIAECILGMNPSRDV (TEL-AML1-319), long anchor-modified TEL-AML1-9V MVSVSPPEEHAMPIGRIAECILGVNPSRDV (TEL-AML1-319-9V) and long anchor-modified gp100 SSGTLISRALVVTHTYLEPGPVTVQVVLQA (gp-265-9V). The long 30-mer peptides were deduced from the natural sequence of each protein. Mice were immunized subcutaneously in the tail base either with 100 nmol (∼ 100 μg) of the nonamer peptides, 50 nmol (∼ 50 μg) of the highly immunogenic decamer peptide Melan-A/MART-1, or 40 nmol (125-130 μg) of the long peptides, diluted in 100 μL PBS with ≤ 10% vol/vol DMSO (Sigma-Aldrich), mixed with 50 μg CpG 1826 (TIB MOLBIOL) and emulsified in 100 μL of incomplete Freund adjuvant (IFA; Sigma-Aldrich), and subsequently boosted (once or twice) with the same peptide emulsions in 2-week intervals, as described.13,21 Pooled splenocytes and inguinal LN cells were analyzed 10-14 days after the boosting. Unspecific peptides used for restimulation in control experiments were Melan-A/MART-1 or NY-BR-1-960 SLSKILDTV (designated NY-BR-1).
In vitro peptide stimulation and intracellular cytokine staining
T-cell function was analyzed as described.13 Spleens and inguinal LNs were isolated from immunized ABabDII mice, prepared as single-cell suspensions and cultured from each mouse separately. Erythrocytes were lysed with ACK lysing buffer (Lonza). Pooled splenocytes and inguinal LN cells were washed 2 times and 1 × 106 cells were incubated overnight with 1μM nonamer peptide (specific or irrelevant) or 25 μL CD3/CD28 Dynabeads (Invitrogen; referred to as anti-CD3/CD28 antibodies). 1 μL/mL GolgiPlug (containing brefeldin A; BD Biosciences) was added 1 hour after the beginning of stimulation. The following day, purified CD16/CD32 antibody was applied to achieve Fc receptor blocking, cells were stained with CD8a FITC, permeabilized and stained with CD3e APC and IFN-γ PE intracellularly, using Cytofix/Cytoperm Plus Kit (BD Biosciences), according to the manufacturer's instruction.
CD107a mobilization assay
Induction of effector CTLs capable of releasing cytolytic granules was measured in the CD107a mobilization assay as described,22,23 with minor modifications. Spleens and inguinal LNs were isolated from immunized ABabDII mice, prepared as single-cell suspensions and cultured from each mouse separately. After lysis of erythrocytes (as described in the previous paragraph), pooled splenocytes and inguinal LN cells were washed 2 times and 1 × 106 cells were incubated for 6 hours with 1μM nonamer peptide (specific or irrelevant peptide) or 25 μL of CD3/CD28 Dynabeads (Invitrogen). PE-conjugated anti-CD107a antibody (BD Biosciences) was present during the whole period of stimulation (at the final concentration 5 μL/mL). 1 μL/mL GolgiStop (containing monensin; BD Biosciences) was added one hour after the beginning of stimulation. Cells were subsequently washed, incubated with purified CD16/CD32 antibody, stained with CD8a FITC and CD3e APC and analyzed by flow cytometry.
Coculture experiments
For peptide loading experiments, NIH-HHD cells were trypsinized, washed and incubated in serum-free medium with 10−5M peptide at room temperature for 4 hours. Afterward, they were washed 3 times, resuspended in the splenocyte medium and 2 × 105 cells were cultured with 1 × 106 pooled splenocytes and LN cells. NIH-HHD-TEL-AML1 and NIH-HHD-Melan-A/MART-1, as well as NIH-HHD not loaded with peptides were plated with splenocytes and LN cells in the same fashion. GolgiPlug was added 1 hour after plating the cells, and T-cell function was analyzed the next day by intracellular cytokine staining as described for peptide stimulation experiments, with different combination of colors (CD8 APC-eFluor 780, CD3e Pacific Blue, and IFNγ PE) to prevent spectral overlapping with EGFP-transduced NIH/3T3 fibroblasts.
T2 cell assay
HLA-A*0201 stabilization assay was done as described,24 with minor modifications. T2 cells (1 × 106/mL) were incubated overnight at 28°C in DMEM with 10% FBS, containing 1 × 10−5M peptide TEL-AML1, TEL-AML1-9V, or Melan-A/MART-1, or left uncoated. The following day, cells were transferred to 37°C, an aliquot was taken at the time point zero, the cells were washed with the medium without FBS and aliquots were taken over a 6-hour period. The aliquots were stained for HLA-A*0201 and MFI was determined by flow cytometry. Fluorescence index (FI) was calculated at the time point zero according to formula FI = MFI [T2 cells with peptide]/MFI [T2 cells without peptide] − 1.
Proteasome purification
Proteasomes were isolated following standard procedures25 from LCL (lymphoblastoid cell lines), which are human B lymphocytes immortalized with EBV mostly expressing active immunoproteasomes. The purity of 20S proteasome preparation was verified by SDS-PAGE electrophoresis, using 12.5% polyacrylamide gel stained with Coomassie dye.
Reverse-phase HPLC and mass spectrometry
In vitro digestion of TEL-AML1-319 peptide and a control peptide listeriolysin O (designated LLO-291; sequence AYISSVAYGRQVYLKLSTNSHSTKVKA) was performed as described.25 1 μg of proteasome was incubated with 10 μg of peptide in a final volume of 100 μL of 20mM HEPES, pH 7.8, 2mM magnesium acetate, 2mM dithiothreitol for the indicated times at 37°C. Digests were stopped with 0.1 volume of trifluoroacetic acid. All digests were repeated at least 3 times.
Peptide analysis and quantification were done as follows: samples were analyzed by reverse-phase HPLC, using an HP1100 system (Hewlett-Packard) equipped with an RPC C2/C18 SC 2.1/10 column (GE Healthcare). Analysis was performed online with an LCQ ion trap MS equipped with an electrospray ion source (ThermoQuest).26 Ion counts of each reaction were normalized to the 9GPS standard peptide, which was added in equal amounts before analysis.25
Results
An anchor modification is needed to render the described TEL-AML1 epitope immunogenic
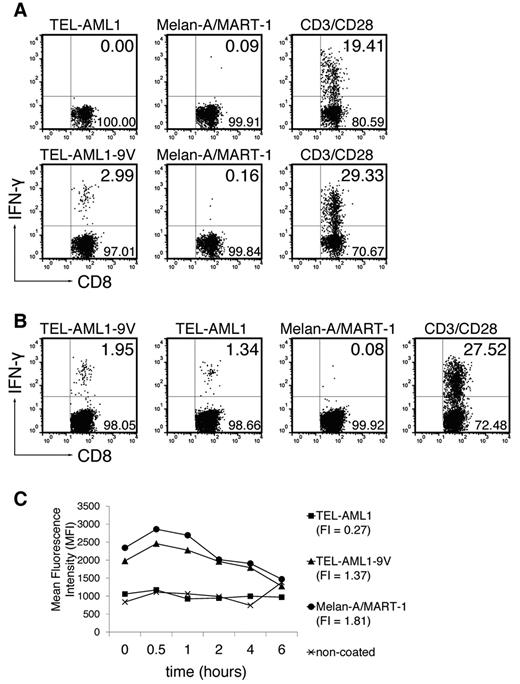
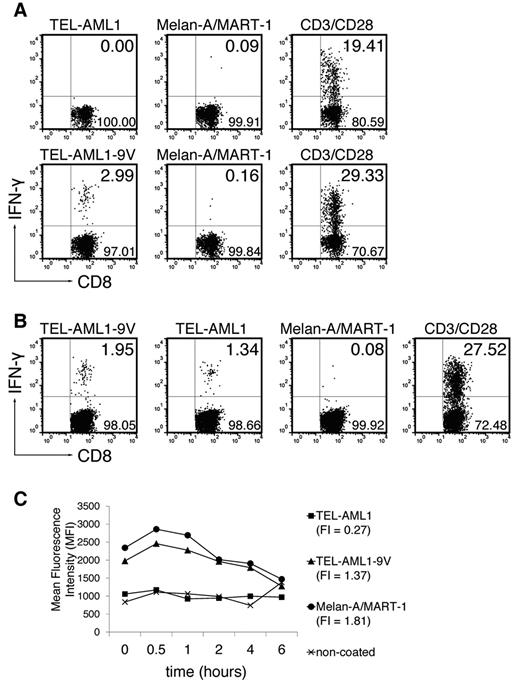
To induce TEL-AML1–specific CD8+ T-cell responses, ABabDII mice were immunized with the exact nonamer peptide epitope (TEL-AML1) in adjuvants (Figure 1A top row) and the specific functional CD8+ T-cell response was tested by intracellular IFN-γ staining, as described.13 Although CD8+ T cells responded to nonspecific polyclonal stimulation through CD3/CD28, no TEL-AML1–specific CD8+ T-cell response could be detected. Therefore, the peptide was anchor-modified by exchanging the C-terminal methionine for valine (TEL-AML1-9V).15 The modification rendered the peptide immunogenic, so that 2.99% of CD3+CD8+ T cells responded specifically to in vitro restimulation with TEL-AML1-9V (Figure 1A bottom row; mean ± SD, 2.64 ± 0.44%), while not responding to restimulation with an irrelevant peptide (anchor-modified Melan-A/MART-1). More importantly, the CD8+ T cells induced in vivo by the anchor-modified peptide specifically recognized the native peptide on in vitro restimulation (Figure 1B; restimulation by the anchor-modified peptide TEL-AML1-9V: mean ± SD, 2.78 ± 0.78%; restimulation by the native TEL-AML1: 1.62 ± 0.25%; restimulation by Melan-A/MART-1: 0.16 ± 0.07%).
TEL-AML1 cannot prime CD8+ T cells unless an anchor modification is introduced, and exhibits very low HLA-A*0201 binding affinity. (A) ABabDII mice were immunized subcutaneously with the native TEL-AML1 peptide (top row) or the anchor-modified TEL-AML1-9V (bottom row), mixed with CpG oligodeoxynucleotides, PBS and IFA. They were boosted twice, and splenocytes and draining LNs were isolated 10-14 days later. The cells were in vitro restimulated overnight with 10−6M corresponding peptide (TEL-AML1 or TEL-AML1-9V), with an irrelevant peptide - anchor-modified Melan-A/MART-1, or with anti-CD3/CD28 antibodies. The cells were stained for CD8, as well as for CD3 and IFN-γ intracellularly, and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants (IFN-γ+ cells residing in the upper right). For the native peptide, 1 representative of 5 analyzed mice is shown; for the anchor-modified, 1 of 3 is shown. (B) ABabDII mice were immunized with the anchor-modified TEL-AML1-9V, and pooled spleen and LN cells were restimulated either with the anchor-modified, the native, the irrelevant peptide or anti-CD3/CD28 antibodies as described in panel A. One representative of 3 mice is shown. (C) T2 cell assay. Cells were either coated with TEL-AML1, TEL-AML1-9V or Melan-A/MART-1 at the final concentration 10−5M, or left uncoated. On the next day, they were stained for HLA-A*0201 and analyzed by flow cytometry. The graph shows changes in MFI during 6 hours and FI of each peptide. One of 2 experiments with similar results is shown.
TEL-AML1 cannot prime CD8+ T cells unless an anchor modification is introduced, and exhibits very low HLA-A*0201 binding affinity. (A) ABabDII mice were immunized subcutaneously with the native TEL-AML1 peptide (top row) or the anchor-modified TEL-AML1-9V (bottom row), mixed with CpG oligodeoxynucleotides, PBS and IFA. They were boosted twice, and splenocytes and draining LNs were isolated 10-14 days later. The cells were in vitro restimulated overnight with 10−6M corresponding peptide (TEL-AML1 or TEL-AML1-9V), with an irrelevant peptide - anchor-modified Melan-A/MART-1, or with anti-CD3/CD28 antibodies. The cells were stained for CD8, as well as for CD3 and IFN-γ intracellularly, and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants (IFN-γ+ cells residing in the upper right). For the native peptide, 1 representative of 5 analyzed mice is shown; for the anchor-modified, 1 of 3 is shown. (B) ABabDII mice were immunized with the anchor-modified TEL-AML1-9V, and pooled spleen and LN cells were restimulated either with the anchor-modified, the native, the irrelevant peptide or anti-CD3/CD28 antibodies as described in panel A. One representative of 3 mice is shown. (C) T2 cell assay. Cells were either coated with TEL-AML1, TEL-AML1-9V or Melan-A/MART-1 at the final concentration 10−5M, or left uncoated. On the next day, they were stained for HLA-A*0201 and analyzed by flow cytometry. The graph shows changes in MFI during 6 hours and FI of each peptide. One of 2 experiments with similar results is shown.
In an attempt to resolve why the native peptide did not evoke specific CD8+ T-cell response, we performed kinetic binding studies using the HLA-A*0201+, transporter associated with antigen processing (TAP)–deficient T2 cell line. Surprisingly, the native peptide virtually did not bind to HLA-A*0201, as opposed to good binding of the anchor-modified one (Figure 1C). In the study by Yotnda et al10 a moderate dissociation rate was reported, with the half-life of complexes of 3 hours, but because the data were not shown, we cannot comment on possible causes of this discrepancy. Nevertheless, taken into account that binding of the native peptide is very weak, it might seem surprising that the CD8+ T cells primed with the anchor-modified peptide could afterward recognize the native one at all. On the other hand, it has been reported that for generating effector CD8+ T cells using heteroclitic peptides, the rate limiting step was the priming step and once the effector cells were generated, the binding affinity of the native peptide to MHC class I was not limiting in the capacity of these cells to perform effector functions.27 The same study reported increased killing of targets expressing native peptides with FI equaling zero. FI of the native TEL-AML1 peptide equals 0.27 (Figure 1C), which might explain its specific recognition by anchor-modified peptide primed CD8+ T cells.
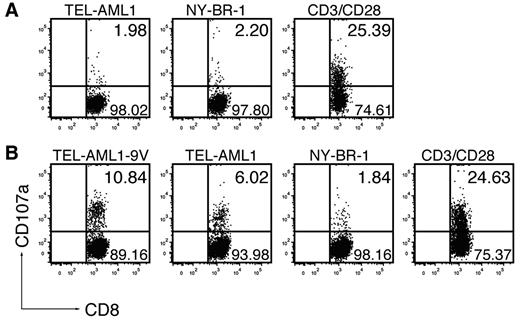
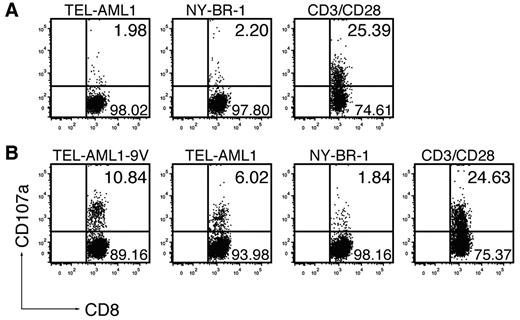
The unexpected absence of immunogenicity of the native TEL-AML1, as opposed to the anchor-modified peptide, prompted us to address this issue by an additional functional assay. We tested whether the peptides could induce cytotoxic T-cell (CTL) effectors, by measuring increase of CD107a (LAMP-1) on CD8+ T-cell surface as a result of degranulation, which directly correlated with their ability to kill.22 Immunization with the native TEL-AML1 peptide (Figure 2A) could not induce specific CTLs in any of the 5 mice tested. On TEL-AML1 peptide restimulation, surface CD107a was detected only in 1.98% of CD8+ T cells (mean ± SD, 2.36 ± 0.79%), which was at the background level, as measured on restimulation with an irrelevant peptide NY-BR-1 (2.2%; mean ± SD, 2.07 ± 0.14%), or without stimulation (data not shown). However, specific CD8+ T cells induced by immunization with the anchor-modified TEL-AML1-9V were bona fide CTLs capable of degranulation, as 10.84% of CD8+ T cells specifically responded to restimulation with TEL-AML1-9V peptide (Figure 2B). Specific responses were seen in all 6 mice tested, ranging from 2.57% to 10.84% (mean ± SD, 5.27 ± 3.13%). As further depicted in Figure 2B, CTLs induced in vivo by the anchor-modified peptide specifically reacted by degranulation on in vitro restimulation with the native TEL-AML1 peptide; specific reactions against the native peptide were seen in 5 of 6 mice, although apparently to a lower extent than against the anchor-modified peptide (ranging from 2.6% to 6.02%; mean ± SD, 3.72 ± 1.27%). After restimulation with the unspecific NY-BR-1, only background levels of CD107a expressing CD8+ T cells were detected (ranging from 1.29% to 2.65%; mean ± SD, 1.88 ± 0.45%). Altogether, functionality of specific CD8+ T-cell effectors, as detected based on IFN-γ production (Figure 1A-B) correlated with their cytotoxic activity (Figure 2), confirming that the native TEL-AML1 was not immunogenic according to any of the criteria applied.
The anchor-modification is needed to induce effector CTLs capable of degranulation. ABabDII mice were immunized subcutaneously with (A) the native peptide TEL-AML1 or (B) the anchor-modified peptide TEL-AML1-9V, mixed with CpG oligodeoxynucleotides, PBS and IFA. They were boosted, and splenocytes and draining LNs were isolated 10 days later. The cells were in vitro restimulated for 6 hours with 10−6M specific peptides (TEL-AML1 or TEL-AML1-9V), with an irrelevant peptide (NY-BR-1), or with anti-CD3/CD28 antibodies, as indicated. Prestaining for CD107a was performed by including anti-CD107a antibodies during the whole period of stimulation, with the addition of monensin. The cells were subsequently stained for CD3 and CD8 and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants. One representative of 5 (A) and 1 of 6 (B) mice are shown, from 2 independent experiments with similar results.
The anchor-modification is needed to induce effector CTLs capable of degranulation. ABabDII mice were immunized subcutaneously with (A) the native peptide TEL-AML1 or (B) the anchor-modified peptide TEL-AML1-9V, mixed with CpG oligodeoxynucleotides, PBS and IFA. They were boosted, and splenocytes and draining LNs were isolated 10 days later. The cells were in vitro restimulated for 6 hours with 10−6M specific peptides (TEL-AML1 or TEL-AML1-9V), with an irrelevant peptide (NY-BR-1), or with anti-CD3/CD28 antibodies, as indicated. Prestaining for CD107a was performed by including anti-CD107a antibodies during the whole period of stimulation, with the addition of monensin. The cells were subsequently stained for CD3 and CD8 and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants. One representative of 5 (A) and 1 of 6 (B) mice are shown, from 2 independent experiments with similar results.
An extended peptide comprising the modified TEL-AML1 epitope cannot induce CD8+ T cells
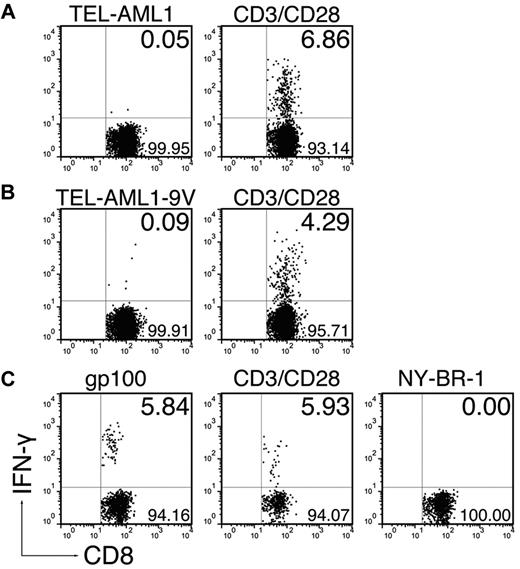
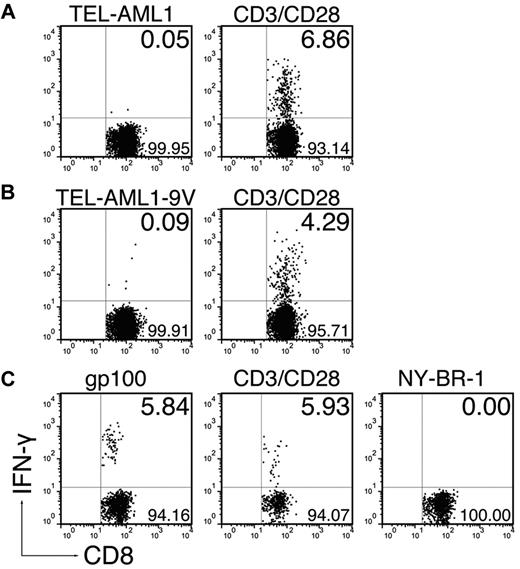
In an attempt to enhance the immune response to the TEL-AML1 peptide, we immunized the mice with extended 30-mer peptides comprising the exact epitope in adjuvants. This immunization strategy has been proven as superior compared with immunization with exact epitopes, because longer peptides are taken up and processed only by professional APCs, which are able to present them in the context of MHC class I molecules.21 Therefore, we were interested to see whether inherent enhanced immunogenicity of long peptides could compensate for the extremely low HLA-A*0201 affinity of the native TEL-AML1 nonamer. We found that the extended native peptide could not evoke a CD8+ T-cell response (Figure 3A, 0.09 ± 0.06%). Surprisingly however, immunization by the 30-mer comprising the immunogenic anchor-modified TEL-AML1-9V nonamer could not induce a CD8+ T-cell response either (Figure 3B; 0.17 ± 0.18%). Because the anchor-modified nonamer could induce the specific response in ABabDII mice, this result strongly suggested that the TEL-AML1 epitope was in fact not endogenously processed. This assumption was supported by the fact that parallel immunization with an extended peptide comprising an anchor-modified epitope derived from human TAA gp100 did induce functional CD8+ T cells, which specifically responded to restimulation in vitro with the exact gp100 epitope (Figure 3C; restimulation by gp100: 3.56 ± 2.07%, restimulation by irrelevant NY-BR-1: 0.07 ± 0.07%).
Elongated peptides comprising the TEL-AML1 epitope cannot evoke CD8+ T-cell responses. ABabDII mice were immunized subcutaneously in the tail base with 30-mer peptides, comprising either (A) the native TEL-AML1 sequence, (B) the anchor-modified TEL-AML1-9V sequence, or (C) the anchor-modified gp100, mixed with CpG oligodeoxynucleotides, PBS, and IFA. In the cases of the TEL-AML1 native and anchor-modified peptide, some of the mice were additionally boosted after 2 weeks with the corresponding peptides. Spleen and draining LNs were isolated 10 days after the immunization, or boosting. The cells were in vitro restimulated overnight either with 10−6M corresponding nonamer peptide (TEL-AML1, TEL-AML1-9V, or gp100), an irrelevant peptide (NY-BR-1) or with anti-CD3/CD28 antibodies. The cells were stained for CD8, as well as for CD3 and IFN-γ intracellularly and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants. In the case of gp100 one representative of 3 analyzed mice is shown, and in the cases of the TEL-AML1 anchor-modified, as well as the native peptide, 1 representative of 6 analyzed mice is shown.
Elongated peptides comprising the TEL-AML1 epitope cannot evoke CD8+ T-cell responses. ABabDII mice were immunized subcutaneously in the tail base with 30-mer peptides, comprising either (A) the native TEL-AML1 sequence, (B) the anchor-modified TEL-AML1-9V sequence, or (C) the anchor-modified gp100, mixed with CpG oligodeoxynucleotides, PBS, and IFA. In the cases of the TEL-AML1 native and anchor-modified peptide, some of the mice were additionally boosted after 2 weeks with the corresponding peptides. Spleen and draining LNs were isolated 10 days after the immunization, or boosting. The cells were in vitro restimulated overnight either with 10−6M corresponding nonamer peptide (TEL-AML1, TEL-AML1-9V, or gp100), an irrelevant peptide (NY-BR-1) or with anti-CD3/CD28 antibodies. The cells were stained for CD8, as well as for CD3 and IFN-γ intracellularly and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants. In the case of gp100 one representative of 3 analyzed mice is shown, and in the cases of the TEL-AML1 anchor-modified, as well as the native peptide, 1 representative of 6 analyzed mice is shown.
The TEL-AML1 peptide is not endogenously processed
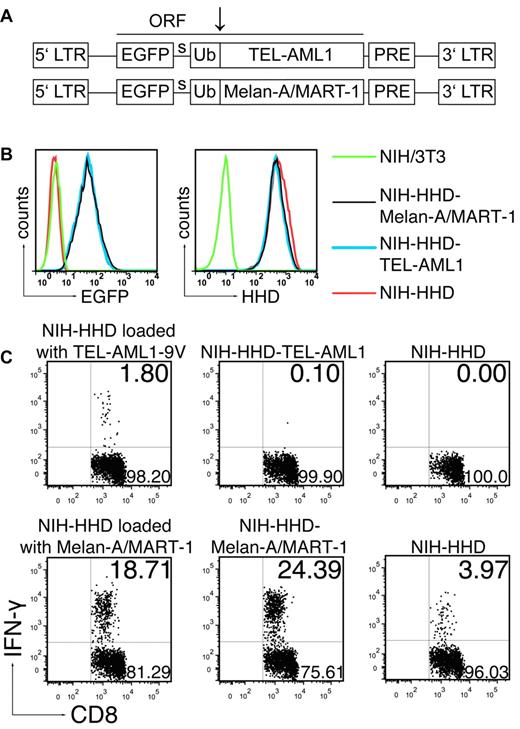
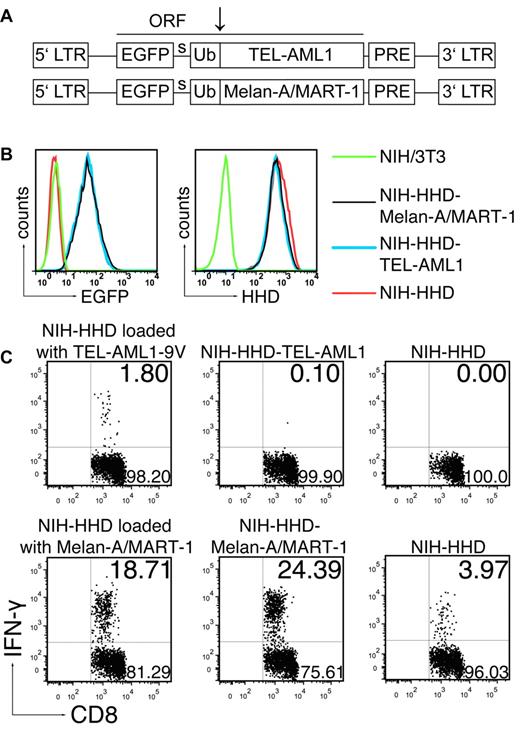
The absence of response to the long anchor-modified TEL-AML1-9V peptide suggested that the described TEL-AML1 epitope was not naturally processed. However, we could not formally exclude that additional factors apart from peptide processing, such as different in vivo peptide degradation kinetics, might have accounted for the response toward gp100 and the absence of such a response to TEL-AML1. Therefore, we tested endogenous processing of the fusion TEL-AML1 region, without influence of other factors affecting success of immunization. To that end, we immunized ABabDII mice with the anchor-modified nonamer peptide TEL-AML1-9V, because we have shown that this mode of immunization induced specific functional CD8+ T cells (Figures 1A-B, 2), and we cocultured the spleen and LN cells with NIH/3T3 fibroblasts transduced to express the HHD restriction element, as well as the minigene encoding the TEL-AML1 fusion region. As an internal control, we generated NIH/3T3 cells expressing HHD and a known human TAA, Melan-A/MART-1. Murine NIH/3T3 fibroblasts were used for generation of HHD-restricted APCs because they were shown to be capable of processing and presenting the same viral and tumor HLA-A*0201 restricted epitopes, which were immunogenic in humans.28,29 Both the TEL-AML1 fusion region and Melan-A/MART-1 were cloned into the retroviral vector pMP7116 in identical fashion, as a single ORF with EGFP and Ub moiety (Figure 4A), according to the Ub/protein/reference (UPR) technique.18 The tripartite fusion comprising the test antigen (TEL-AML1), or the control tumor antigen (Melan-A/MART-1) is cleaved cotranslationally, or nearly so, by Ub-specific proteases, producing equimolar amounts of the protein of interest (TEL-AML1 or Melan-A/MART-1) and the reference protein (EGFP), thus enabling precise comparison of expression levels of the test and the control antigen. The levels of expression of TEL-AML1 and Melan-A/MART-1, measured as EGFP fluorescence intensity by flow cytometry, were almost identical in both cell lines (NIH-HHD-TEL-AML1 and NIH-HHD-Melan-A/MART-1; Figure 4B left). The levels of HHD expression were also almost identical in both cell lines, as well as the level in the NIH/3T3 cells transduced to express only HHD (NIH-HHD; Figure 4B right).
The TEL-AML1 peptide is not naturally processed. (A) Schematic representation of the 2 MP71 constructs. EGFP, ubiquitin (Ub) and either TEL-AML1 minigene or Melan-A/MART-1 constitute a single ORF. Site of cleavage by Ub-specific proteases is indicated by arrow. LTR indicates long terminal repeat; PRE, woodchuck hepatitis virus posttranscriptional regulatory element; and s, spacer peptide. (B) Levels of expression of TEL-AML1 and Melan-A/MART-1, measured by EGFP expression (left), and of HHD, measured after staining with HLA-A*0201 specific antibody (right) in the transduced cell lines. (C) ABabDII mice were immunized either with TEL-AML1-9V (top row) or Melan-A/MART-1 peptide (bottom row) as described in Figure 1 and boosted after 14 days. Ten days after the boost spleen and LNs were isolated, pooled and cocultured overnight either with the NIH-HHD cells loaded with the corresponding peptide, or with NIH-HHD endogenously expressing TEL-AML1 fusion region (NIH-HHD-TEL-AML1) or Melan-A/MART-1 (NIH-HHD-Melan-A/MART-1). In addition, they were cultured with NIH-HHD cells only. On the next day, the cells were stained for CD8, as well as for CD3 and IFN-γ intracellularly, and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants. In each case one representative of 3 analyzed mice is shown.
The TEL-AML1 peptide is not naturally processed. (A) Schematic representation of the 2 MP71 constructs. EGFP, ubiquitin (Ub) and either TEL-AML1 minigene or Melan-A/MART-1 constitute a single ORF. Site of cleavage by Ub-specific proteases is indicated by arrow. LTR indicates long terminal repeat; PRE, woodchuck hepatitis virus posttranscriptional regulatory element; and s, spacer peptide. (B) Levels of expression of TEL-AML1 and Melan-A/MART-1, measured by EGFP expression (left), and of HHD, measured after staining with HLA-A*0201 specific antibody (right) in the transduced cell lines. (C) ABabDII mice were immunized either with TEL-AML1-9V (top row) or Melan-A/MART-1 peptide (bottom row) as described in Figure 1 and boosted after 14 days. Ten days after the boost spleen and LNs were isolated, pooled and cocultured overnight either with the NIH-HHD cells loaded with the corresponding peptide, or with NIH-HHD endogenously expressing TEL-AML1 fusion region (NIH-HHD-TEL-AML1) or Melan-A/MART-1 (NIH-HHD-Melan-A/MART-1). In addition, they were cultured with NIH-HHD cells only. On the next day, the cells were stained for CD8, as well as for CD3 and IFN-γ intracellularly, and analyzed by flow cytometry. Events shown are gated on CD3+CD8+ lymphocytes; numbers indicate percentages of cells in the respective quadrants. In each case one representative of 3 analyzed mice is shown.
To show that the generated NIH-HHD cells can efficiently present peptide antigens to CD8+ T cells, we loaded the cells with the nonamer peptides used for the immunization (TEL-AML1-9V or Melan-A/MART-1) and cocultured them with the pooled spleen and LN cells from the immunized mice. After overnight coculture, CD8+ T cells specifically responded to the corresponding peptide used for immunization (Figure 4C), giving 1.8% (mean ± SD, 1.87 ± 0.73%) of IFN-γ+ CD3+CD8+ T cells in the case of TEL-AML1-9V and 18.71% (mean ± SD, 24.85 ± 5.66%) in the case of Melan-A/MART-1. The response was specific, because stimulation only with the NIH-HHD cells gave 0% IFN-γ+ cells in the case of TEL1-AML1-9V, and only 3.97% in the case of highly immunogenic Melan-A/MART-1 (mean ± SD, 0.1 ± 0.14% for TEL-AML1-9V, 6.81 ± 2.48% for Melan-A/MART-1). The slight background seen in the case of Melan-A/MART-1 was likely because of retention of this high-affinity binding peptide on APCs present in spleen, because it was abolished when the CD8+ cells were additionally magnetically negatively sorted (data not shown). However, the induced CD8+ T cells in the TEL-AML1-9V immunized mice were not able to recognize NIH/3T3 fibroblasts expressing TEL-AML1 endogenously (Figure 4C; 0.1% IFN-γ+ T cells; mean ± SD, 0.15 ± 0.05%). On the contrary, CD8+ T cells from Melan-A/MART-1 immunized mice responded very well to stimulation with NIH cells expressing Melan-A/MART-1 endogenously (24.39% IFNγ+ T cells; mean ± SD, 32.56 ± 7.47%). The response was even slightly stronger than the response to NIH-HHD cells exogenously loaded with the peptide, arguing that the NIH-HHD cells not only efficiently presented peptides to T cells, but also endogenously processed the UPR-derived polypeptides even more efficiently. Therefore, if the TEL-AML1 fusion region would have been processed, we would have been able to detect it, most probably as a stronger response in comparison to the one seen with the exogenously added peptide.
The TEL-AML1 peptide is not processed by human proteasomes in vitro
There is substantial evidence suggesting a large functional redundancy between murine and human antigen processing machineries in general, and particularly with regard to HLA class I restricted human epitopes.17,30-36 Moreover, our results confirm that the murine antigen processing machinery correctly generates the same tumor-derived epitopes, which are processed in humans—both in ABabDII mice (gp100 epitope, Figure 3C) and in murine NIH/3T3 fibroblasts (Melan-A/MART-1 epitope, Figure 4C). However, we wanted to confirm that the TEL-AML1 epitope could not be processed by human proteasomes. To that end, the 30 amino acid long peptide spanning the TEL-AML1 fusion region (TEL-AML1-319) was digested by purified lymphoblastoid cell lines (LCL) 20S proteasomes. Peptide fragments were detected by reverse-phase liquid chromatography coupled online with tandem mass spectrometry (LC-MS/MS; Figure 5A). The described epitope was not identified. Moreover, longer fragments with the appropriate C-terminus, which could serve as potential precursors for amino-terminal trimming by aminopeptidases present in the cytosol and ER were not identified either. Instead, kinetic analysis identified a large number of destructive cleavages within the epitope, which formed relatively early during the digestion and destroyed the epitope. Control digestion of a polypeptide derived from bacterial listeriolysin O (LLO-291) resulted in the efficient generation of both the MHC class I epitope LLO-296-304, as well as its N-terminally elongated precursor (Figure 5B).
In vitro digestion of synthetic peptides with human proteasomes. (A) Digestion of the peptide TEL-AML1-319. Predominant cleavage products and cleavage sites are shown. Synthetic peptides were incubated until 50% of the peptide substrate was turned over by purified LCL (lymphoblastoid cell lines) 20S proteasomes, which mainly expresses immunoproteasomes. Peptide fragments are detected by LC-MS/MS (reversed phase liquid chromatography coupled online with tandem mass spectrometry) using a normal triple method. Arrows of various thickness are proportional to the relative cleavage intensity. Note the large number of destructive cleavages within the epitope (in red) and the complete absence of a potentially functional N-terminal cleavage. (B) Generation of the LLO-296-304 CD8+ T-cell epitope by digestion of synthetic polypeptide LLO-291 derived from bacterial listeriolysin O. The experiment was performed as described in panel A. The dominant relevant cleavage products and the MHC class I ligand Val296-Leu304 (in red) as well as the N-terminally elongated epitope precursor peptide thereof (in green) are shown. Arrows indicate major and minor cleavage sites. Note the strong cleavage behind the C-terminal Leu304 residue, resulting in the predominant generation of the epitope. Cleavage within the epitope does not effect the overall predominance of Val296-Leu304 peptide generation. Numbers designate amino acid residue positions in the corresponding proteins (TEL-AML1 and listeriolysin O).
In vitro digestion of synthetic peptides with human proteasomes. (A) Digestion of the peptide TEL-AML1-319. Predominant cleavage products and cleavage sites are shown. Synthetic peptides were incubated until 50% of the peptide substrate was turned over by purified LCL (lymphoblastoid cell lines) 20S proteasomes, which mainly expresses immunoproteasomes. Peptide fragments are detected by LC-MS/MS (reversed phase liquid chromatography coupled online with tandem mass spectrometry) using a normal triple method. Arrows of various thickness are proportional to the relative cleavage intensity. Note the large number of destructive cleavages within the epitope (in red) and the complete absence of a potentially functional N-terminal cleavage. (B) Generation of the LLO-296-304 CD8+ T-cell epitope by digestion of synthetic polypeptide LLO-291 derived from bacterial listeriolysin O. The experiment was performed as described in panel A. The dominant relevant cleavage products and the MHC class I ligand Val296-Leu304 (in red) as well as the N-terminally elongated epitope precursor peptide thereof (in green) are shown. Arrows indicate major and minor cleavage sites. Note the strong cleavage behind the C-terminal Leu304 residue, resulting in the predominant generation of the epitope. Cleavage within the epitope does not effect the overall predominance of Val296-Leu304 peptide generation. Numbers designate amino acid residue positions in the corresponding proteins (TEL-AML1 and listeriolysin O).
Discussion
In the present study, we have dissected applicability of a potentially very attractive target epitope for T cell–mediated therapy of childhood BCP-ALL. The described TEL-AML1 epitope could not prime immune responses in ABabDII mice, most probably because of its low HLA-A*0201 binding affinity. Modification of an anchor residue was needed to render the peptide immunogenic. Effector CD8+ T cells primed in vivo with the anchor-modified peptide retained the specificity for the native TEL-AML1, as they specifically responded to in vitro restimulation with the native peptide by IFN-γ secretion and by degranulation, although this cytolytic response was apparently lower. Even though we succeeded in inducing specific CD8+ T-cell responses against the TEL-AML1 peptide, we presented multiple evidence showing that this peptide could not be correctly processed. After long peptide immunization, human TAA gp100 epitope was correctly processed in vivo in ABabDII mice leading to specific T-cell response induction, whereas extended peptide comprising the immunogenic anchor-modified TEL-AML1 variant repeatedly failed to evoke such response. An approach designed to directly address the issue of processing, whereby potent APCs were generated and shown to efficiently process and present another human TAA Melan-A/MART-1, revealed that the TEL-AML1 peptide was not endogenously processed. Importantly, the model system used enabled precise quantitative comparison of the expression levels of the control Melan-A/MART-1 and the test TEL-AML1 antigen in the APCs. The levels were virtually identical, hence could not account for the observed results. Finally, even when human proteasomes purified from immortalized B lymphocytes were used, the TEL-AML1 peptide could not be generated, although the control digestion resulted in efficient production of both the listeriolysin O epitope and its N-terminally extended precursor.
The proteasome consists of different subunits and their abundance within the cell determines predominance of certain proteasomal forms, such as constitutive and immunoproteasome.37 Direct comparison of peptide fragments generated by constitutive proteasomes isolated from mouse C4 fibroblasts and human carcinoma HeLa cells, as well as such comparison involving immunoproteasomes isolated from the same murine and human cells after IFN-γ treatment, revealed no species-specific proteasomal cleavage properties.38 Immunoproteasomes compared with constitutive proteasomes, however, do exhibit altered cleavage site preferences, and therefore generate certain epitopes with very different efficiencies.39 Nevertheless, direct mass spectrometric analyses of digested polypeptides comprising such epitopes revealed large, yet solely quantitative differences in the epitopes (or epitope precursors) generated by constitutive, compared with immunoproteasomes.40-43 Human LCL-derived proteasomes used here for digestion analysis are predominantly immunoproteasomes. Because we do not know the precise composition of proteasomes which might be expressed in leukemic blasts in vivo, we cannot completely exclude that in these cells the TEL-AML1 peptide might be generated. However, in view of the studies discussed earlier in this paragraph,40-43 and taking into account multiple destructive cleavages within the proposed TEL-AML1 epitope detected, that should be unlikely. Furthermore, mere avoidance of the complete destruction might not necessarily allow this peptide to act as a tumor rejection antigen. One might hypothesize that the TEL-AML1 peptide would have to be generated very efficiently, to allow appearance of a sufficient number of surface TEL-AML1/MHC class I complexes, to sustain an effective T-cell response leading to complete eradication of leukemia-propagating cells. Potential requirement for a high level of processing seems justified, as TEL-AML1 exerts indeed a very poor ability of stabilizing MHC class I heterodimeric complexes and high peptide amounts might be therefore needed to override dissociation of the unstable complexes that would form. By the same token, although it cannot be excluded that leukemic blasts in vivo might acquire a proteasomal processing defect, which would allow the TEL-AML1 peptide to appear at the cell surface in MHC class I peptide binding cleft at very low levels, relying on such a putative deficiency in achieving complete therapeutic effect might be questionable.
Our results are in contrast with those from the study by Yotnda et al.10 In their study, the TEL-AML1 peptide was used to prime CTLs, which were reported to specifically recognize autologous leukemic cells and the REH cell line. However, most of the cytotoxicity assays in the study were performed with T-cell lines, not with isolated T-cell clones. Because T-cell specificity is inherently cross-reactive,44 a polyclonal population can be expected to respond nonspecifically to some extent. In the aforementioned study, cytotoxic activities of the T-cell line and the isolated CTL clones were low, with high levels of background killing, making it difficult to judge whether the reactivity was indeed specific for the TEL-AML1 fusion peptide. Moreover, comparable (and low) levels of killing of both HLA-A*0201-transfected and the nontransfected REH line pose a similar question regarding HLA-A*0201 restriction.
In conclusion, although the described TEL-AML1 fusion peptide virtually does not bind to HLA-A*0201, this problem could be overcome by modifying the anchor-residue, the strategy which has been and is applicable to a variety of peptides proposed for immunotherapy.45 However, as our study reveals the lack of natural processing of the TEL-AML1 peptide, we conclude that it cannot be proposed as a potential target in cancer immunotherapy in HLA-A*0201-restricted fashion. Although true tumor-specific antigens might indeed present the most attractive targets, these findings highlight the need for a careful evaluation of proposed epitopes, because only efficiently processed and presented peptides would be suited for therapy.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Laurence Chapatte for providing the lentiviral vectors pRRL.CMV.GFP.sin18; Christel Westen, Marion Rösch, Angelika Gärtner, Stephanie Fürl, and Christin Keller for technical assistance; Hans-Peter Rahn for FACS sorting; Daniel Sommermeyer for discussions and critically reading the manuscript; and Jehad Charo for discussions.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG)–Sonderforschungsbereich TR36 and the Helmholtz-Gemelnschaft Deutscher Forshcuncszentren (HA-202). P.M.K. was supported through DFG project KL 427/15-1.
Authorship
Contribution: J.P. designed and performed experiments, interpreted data, and wrote the manuscript; L.L.P. generated and provided mice; P.M.K. designed and performed experiments; M.L. and W.U. provided HHD expression vector; and T.B. designed research, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Blankenstein, Max-Delbrück-Centrum für Molekulare Medizin, Robert-Rössle-Str 10, 13092 Berlin, Germany; e-mail: tblanke@mdc-berlin.de.