Abstract
The deregulation of translation markedly contributes to the malignant phenotype in cancers, and the assembly of the translation initiating complex eIF4F is the limiting step of this process. The mammalian Target of Rapamycin Complex 1 (mTORC1) is thought to positively regulate eIF4F assembly and subsequent oncogenic protein synthesis through 4E-BP1 phosphorylation. We showed here that the translation inhibitor 4EGI-1 decreased the clonogenic growth of leukemic progenitors and induced apoptosis of blast cells, with limited toxicity against normal hematopoiesis, which emphasize the importance of translation deregulation in acute myeloid leukemia (AML) biology. However, the mTORC1 inhibitor RAD001 (a rapamycin derivate) did not induce AML blast cell apoptosis. We herein demonstrated that mTORC1 disruption using raptor siRNA or RAD001 failed to inhibit 4E-BP1 phosphorylation in AML. Moreover, RAD001 failed to inhibit eIF4F assembly, to decrease the proportion of polysome-bound c-Myc mRNA, and to reduce the translation-dependent accumulation of oncogenic proteins. We identified the Pim-2 serine/threonine kinase as mainly responsible for 4E-BP1 phosphorylation on the S65 residue and subsequent translation control in AML. Our results strongly implicate an mTORC1-independent deregulation of oncogenic proteins synthesis in human myeloid leukemogenesis. Direct inhibition of the translation initiating complex thus represents an attractive option for the development of new therapies in AML.
Introduction
Acute myeloid leukemia (AML) is a clonal hematologic disease characterized by differentiation arrest and by the proliferation of immature myeloid progenitors, both sustained by the deregulation of multiple signaling pathways.1 Despite recent advances in the understanding of AML biology, the prognosis of this disease remains poor and new therapeutic perspectives are therefore under active investigation.2
In most cancer models, the activity of mammalian Target of Rapamycin Complex 1 (mTORC1) is dependent on the activation of Akt, an oncoprotein that operates downstream of class IA phosphoinositide 3-kinase (PI3K).3 In AML, mTORC1 is frequently activated,4 but recent evidence has suggested that this activity does not rely on PI3K/Akt. These findings have shown that the p110δ isoform of PI3K is principally responsible for PI3K activity5 and that a specific p110δ inhibitor, IC87114, fully inhibits Akt phosphorylation without affecting mTORC1 activity.6 Moreover, the Src kinase Lyn has recently been shown to control mTORC1 activity but not Akt phosphorylation in primary AML cells.7
The mTORC1 complex consists of mTOR, raptor, mLST8, and PRAS40.8 It governs cell growth and regulates the cap-dependent translation of mRNAs.9 Rapamycin and derivates, referred to as rapalogs (eg, RAD001), are closely related molecules demonstrating similar biologic activities by specifically repressing mTORC1 activity.10,11 These compounds have recently been developed as anticancer therapeutics because of the frequent activation of mTORC1 in cancers and approved for clinical use.12 Despite their seemingly clear mechanism of action and the rationale for their use as a cancer therapeutic, rapalogs have had only modest successes in clinical trials.3 In the case of AML, mTORC1 is frequently activated and rapamycin reduces the clonogenic growth of AML cells and sensitizes blast cells to chemotherapy-induced cell death.4 However, rapamycin barely induces apoptosis and only after long-term exposure.4 Accordingly, the clinical efficacy of rapamycin in monotherapy is limited in AML.13
Different mechanisms of resistance to rapamycin have been described in human cancer cell lines.14 In primary AML cells, mTORC1 inhibition using RAD001 increases PI3K/Akt activity via an insulin-like growth factor-1 autocrine loop.6 However, this mechanism provides only a partial explanation of resistance to rapamycin as the dual inhibition of mTORC1 using RAD001 and PI3K using IC87114 reduces AML blast cell proliferation but does not induce apoptosis.6,13,15
As the mTORC1 pathway is known to stimulate protein synthesis, the moderate antileukemic activity of rapamycin led us to investigate the importance of translation in AML biology. In eukaryotic cells, translation is tightly regulated at the initiation level and is controlled by the mTORC1 pathway. The serine/threonine kinase activity of mTORC1 is dependent on raptor, which positively regulates mTOR by acting as a scaffold for mTORC1 substrates, the ribosomal protein S6 kinase (S6K), and the eukaryotic initiation factor 4E (eIF4E)–binding proteins (4E-BPs).3,16,17 The 4E-BPs are necessary for the control of translation initiation downstream of mTORC1 and although 3 isoforms exist in mammals, most studies have focused on 4E-BP1. The eIF4E protein recognizes the 7-methyl-GTP (7m-GTP) cap structure at the 5′ end of mRNA molecules18 and associates with the scaffold protein eIF4G to form the eIF4F translation initiation complex. 4E-BP1 inhibits the formation of eIF4F by competitively inhibiting the recruitment of eIF4G and sequestering eIF4E in an inactive complex. 4E-BP1 can be sequentially phosphorylated on multiple residues. Primary phosphorylations on T37 and T46 residues are dependent of mTOR, although they do not require raptor and are somewhat insensitive to rapamycin.19,20 They are necessary for subsequent phosphorylation on T70 and then on S65 that are critical for the release of 4E-BP1 from eIF4E.21-23
In contrast to the observations made in AML, this model predicts a high degree of antitumor activity using mTORC1 inhibitors. Indeed, mechanisms implicated in protein synthesis strongly contribute to malignancy. A direct deregulation of translation initiation by eIF4E overexpression or by constitutive 4E-BP1 phosphorylation is commonly found in cancers and contributes to the oncogenic process through a sustained translation of malignancy-related transcripts, such as c-Myc and Cyclin D1.24 Indeed, mRNAs are translated with various efficiencies depending on the structure of their 5′untranslated region (UTR). The short unstructured 5′ends of mRNAs encoded by housekeeping genes (ie, “strong” mRNAs: actin) enable efficient translation under basal conditions. Conversely, the long structured 5′UTR of mRNA molecules encoded by oncogenes (“weak” mRNAs: c-Myc, Cyclin D1, and Bcl-xL) require an increased availability of eIF4F to achieve efficient translation.18,25,26
We show herein that targeting translation demonstrated a marked antileukemic potential in AML. In primary AML cells, specific inhibition of cap-dependent translation was achieved using a 4E-BP1 mimetic, the small molecule 4EGI-1.27 This compound dramatically decreased the clonogenic growth of AML precursors and induced massive blast cell apoptosis, with minimal impairment of normal hematopoiesis ex vivo. However, mRNA translation escapes mTORC1 control in primary AML cells. The phosphorylation of P70S6K on T389, a typical downstream relay of mTORC1, was abrogated by RAD001 or raptor siRNA, whereas the phosphorylation of 4E-BP1 on S65 residue, essential for the control of translation initiation, was unaffected. Therefore, RAD001 failed to inhibit mRNA translation: unlike 4EGI-1, RAD001 did not inhibit the assembly of eIF4F complexes and did not reduce the association of c-Myc mRNA with polysomes. We identified the Pim-2 serine/threonine kinase, acting in a raptor-independent and rapamycin-insensitive way, as mainly responsible for 4E-BP1 phosphorylation on S65 and subsequent cap-dependent translation control in AML. Indeed, pim-2 knock-down and treatment of AML cells by 4EGI-1 strongly reduced the accumulation of oncogenic proteins regulated at the translation initiation level (eg, c-Myc, Cyclin D1, Bcl-xL), whereas RAD001 failed to modify their cellular levels.
Our results implicate a rapamycin-insensitive deregulation of oncogenic protein expression in human myeloid leukemogenesis and strongly support the development of therapeutic strategies that directly target the eIF4F translation initiating complex in AML.
Methods
Patients
Bone marrow samples were obtained from 24 patients with newly diagnosed AML; all included in chemotherapy trials initiated by the French Multicenter Group, Groupe Ouest Est des Leucémies et Autres Maladies du Sang (GOELAMS). All studies were approved by the GOELAMS Institutional Review Board, and signed informed consent was obtained in accordance with the Declaration of Helsinki.
Cells, cultures, and reagents
Blast cells were isolated from bone marrow aspirates by Ficoll-Hypaque gradient density centrifugation as previously described.5 Only samples with more than 80% of blast cells were used. Normal peripheral CD34+ cells were purified from 6 healthy donors after informed consent using MIDI-MACS immunoaffinity columns (Miltenyi Biotec; > 95% CD34+ cells). LY294002 was purchased from Sigma-Aldrich, RAD001 was provided by Novartis, and 4EGI-1 was kindly given by Dr Gerhard Wagner (Harvard Medical School, Boston, MA).
Colony assays
Clonogenic assays of normal CD34+ hematopoietic progenitors.
Normal CD34+ cells were plated in duplicate (5 × 104/mL) in 1 mL H4100 medium (StemCell Technologies) with 10% fetal calf serum (FCS), 0,8% bovine serum albumin, 1.7 mM glutamine, 10−4 mol/L β-mercaptoethanol), 1 IU/mL erythropoietin, 2.5 ng/mL granulocyte-macrophage colony-stimulating factor, 1000 IU/mL IL-3, 10 ng/mL stem cell factor, 20 ng/mL IL-6, and 2.5 ng/mL granulocyte colony-stimulating factor. The erythroid (BFU-E), granulo-macrophagic (CFU-GM), and granulo-erythroid–megakaryocytic-monocytic (CFU-GEMM) colony-forming units were counted under an inverted microscope.28
CFU-L assays.
Flow cytometry
Apoptosis was quantified by annexin V binding. A total of 2 × 105 cells were cultured without or with 4EGI-1 during 24 or 48 hours and then labeled with annexin V–PE (BD Biosciences) according to the manufacturer's instructions.
RNAi
SiRNA targeting human Pim-2 (Ambion), human raptor (sc-44 069; Santa Cruz Biotechnology), and control nontargeted siRNAs (Dharmacon RNA Technologies) were transfected using the Amaxa nucleofector following the manufacturer's instruction (Amaxa Biosystems); 2 × 106 blast cells were resuspended in 100 μL of nucleofector solution L (VCA-1005). Then 1-μM oligonucleotides were added. After electroporation (X-001 program), cells were placed in 2 mL of 10% FCS minimum essential medium (MEM) and analyzed by Western blot after 24 hours.
7-Methyl-guanosine cap affinity assay
A total of 5 × 106 cells were lysed by 3 freeze-thaw cycles in 600 μL of solubilization buffer (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid/KOH (HEPES/KOH), pH 7.6, 200 mM KCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 20 mM KF, 1 mM K4P2O7, 10% glycerol, 1% NP40, protease inhibitors [Roche Diagnostics] and 50 μg/mL RNAse-A). Cell lysates were clarified by centrifugation (13 000g, 20 minutes, 4°C), and supernatants were incubated (2 hours, 4°C) with 7m-GTP-Sepharose beads (GE Healthcare) in 400 μL binding buffer (50 mM Tris/HCl, pH 7.5, 300 mM KCl, 1 mM EDTA, 20 mM KF, 1 mM K4P2O7, 1 mM dithiothreitol (DTT), and protease inhibitors). Beads were then washed 3 times in binding buffer and boiled in Laemmli sample buffer.
Polysome analysis
MOLM-14 cells pellets were lysed in hypotonic buffer (50 mM Tris/HCl, pH 7.5, 7 mM MgCl2, 100 mM KCl, 2 mM DTT, 1% NP-40, 500 IU/mL RNAsin) and clarified by centrifugation (13 000g, 2 minutes, 4°C). Supernatants were loaded onto 10% to 50% sucrose density gradients in 20 mM HEPES/KOH (pH 7.5), 100 mM KCl, 2 mM MgCl2, and centrifuged (Beckman SW41Ti rotor, 35 000g, 2 hours, 4°C). After centrifugation, 12 fractions (1 mL) were collected using a Beckman fraction recovery system. To purify RNA, 1% sodium dodecyl sulfate and 50 mg/mL proteinase K were added and samples were incubated (30 minutes, 37°C). One volume of phenol/chloroform/isoamylalcohol (25:24:1) was added, and the samples were centrifuged (10 minutes, 16 000g). RNA was precipitated with 0.2 volume of 7.5 M ammonium acetate and 2 volumes of absolute ethanol. RNA pellets were washed with 70% ethanol and solubilized in water.
[3H]Leucine incorporation assay
Blast cells were incubated for 2 hours, at 105/mL in a low leucine medium (90% Dulbecco modified Eagle medium without leucine, 10% αMEM, with 10% dialysed FCS), without or with inhibitors, then labeled for 1 hour with [3H]leucine (1 μCi, 37kBq). The amount of radioactivity incorporated into proteins was determined by trichloroacetic acid precipitation.
Western blot
Cells were boiled in Laemmli sample buffer, and proteins from 106 cells were resolved by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with primary antibodies. Anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti–phospho-BAD S118, antiraptor, anti-mTOR, anti–Pim-2, anti–4E-BP1, anti-eIF4E, anti-eIF4G, and anti–caspase-3 antibodies were from Cell Signaling Technology; anti-P70S6K, anti-BAD, anti–c-Myc, anti–Cyclin D1, and anti–Bcl-xL antibodies were from Santa Cruz Biotechnology, and antiactin was from Sigma-Aldrich. Proteins were visualized using a secondary antibody conjugated to horseradish peroxidase and chemiluminescence detection (enhanced chemiluminescence, GE Healthcare). The images were captured using a CCD camera (LAS3000 from FujiFilm) and quantified using Multigauge software from Fujifilm.
Real-time quantitative PCR
Reverse transcription and real-time polymerase chain reaction (PCR) were performed as previously described.29 Primers for human c-Myc were as follows: forward: GGAACTCTTGTGCGTAAGGA; reverse: CAAGACTCAGCCAAGGTTGTG. Primers for human β-actin were as follows: forward: TCCCTGGAGAAGAGCTACGA; reverse: AGCACTGTGTTGGCGTACAG.
Statistical analysis
Statistical significance of differences observed between experimental groups was determined using Student t test. In each case using primary AML cells, independent experiments mean that the experiments were performed in different days and that cells from different patients were used.
Results
4EGI-1 inhibits the clonogenic growth of AML progenitors and induces their apoptosis with minimal impairment of normal hematopoiesis
The 4EGI-1 compound has recently been identified through a high-throughput screening assay for small-molecule inhibitors of the eIF4E/eIF4G interaction.27 Thus, 4EGI-1 behaves as a cell-permeable 4E-BP1 mimetic that inhibits cap-dependent mRNA translation. This compound exhibits growth inhibitory activity against multiple cancer cell types at concentrations ranging from 50 to 200 μM.27
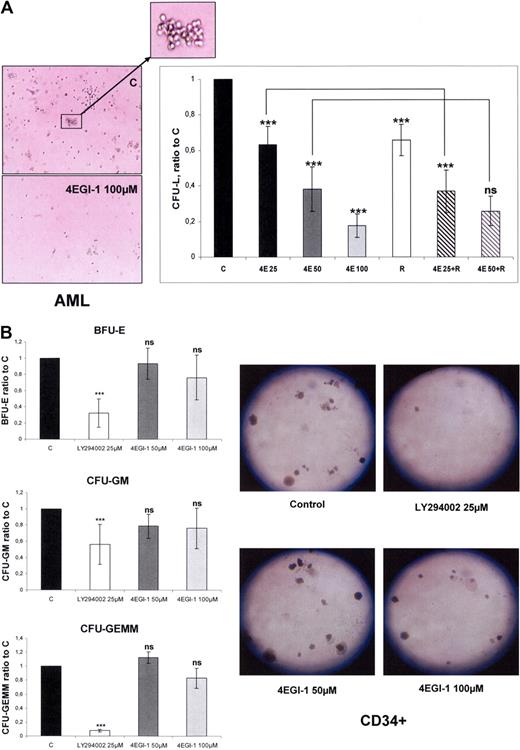
We performed clonogenic cultures of primary AML cells incubated with increasing concentrations of 4EGI-1 and/or 10 nmol/L RAD001. Colony formation by leukemic progenitors from 5 different AML samples was dramatically reduced by 4EGI-1 with a dose-dependent response (mean decrease of 37%, 62%, and 83% with 4EGI-1 25, 50, and 100 μM, respectively; Figure 1A). Furthermore, incubation of AML cells with 10 nmol/L RAD001 reduced the number of CFU-L, as reported using rapamycin,4 and an additive effect of suboptimal concentrations of 4EGI-1 (25 μM) was noted (Figure 1A). These results suggested that 4EGI-1 and RAD001 may have reduced CFU-L by repressing distinct cellular processes.
4EGI-1 dramatically reduces the clonogenic growth of AML progenitors with a moderate impairment of normal CD34+ hematopoietic progenitor clonogenicity. (A) AML blast cells from 5 patients were plated in H4230 methylcellulose medium without or with 25, 50, or 100 μM 4EGI-1 (4E25, 4E50, and 4E100, respectively), 10 nmol/L RAD001 (R), or the association of RAD001 with 25 μM or 50 μM 4EGI-1. Cells were then incubated for 7 days, and the leukemic colonies (> 20 cells), referred to as CFU-L, were scored under an inverted microscope. In the control condition, the numbers of CFU-L were 66 to 258 in 5 independent experiments. (B) Normal CD34+ cells were purified from 5 healthy donors and then plated as described in “Colony assays,” without or with 50 or 100 μM 4EGI-1. The BFU-E, CFU-GM, and CFU-GEMM colony-forming units were counted under an inverted microscope. In the control condition, the numbers of colonies were 82 to 118, 43 to 58, and 11 to 15 for BFU-E, CFU-GM, and CFU-GEMM, respectively, in 5 experiments. Each histogram represents the mean of 5 independent experiments, performed in duplicates. Results are expressed as a ratio between each condition and the control condition. Vertical bars represent SD. ***P < .001. ns indicates not significant.
4EGI-1 dramatically reduces the clonogenic growth of AML progenitors with a moderate impairment of normal CD34+ hematopoietic progenitor clonogenicity. (A) AML blast cells from 5 patients were plated in H4230 methylcellulose medium without or with 25, 50, or 100 μM 4EGI-1 (4E25, 4E50, and 4E100, respectively), 10 nmol/L RAD001 (R), or the association of RAD001 with 25 μM or 50 μM 4EGI-1. Cells were then incubated for 7 days, and the leukemic colonies (> 20 cells), referred to as CFU-L, were scored under an inverted microscope. In the control condition, the numbers of CFU-L were 66 to 258 in 5 independent experiments. (B) Normal CD34+ cells were purified from 5 healthy donors and then plated as described in “Colony assays,” without or with 50 or 100 μM 4EGI-1. The BFU-E, CFU-GM, and CFU-GEMM colony-forming units were counted under an inverted microscope. In the control condition, the numbers of colonies were 82 to 118, 43 to 58, and 11 to 15 for BFU-E, CFU-GM, and CFU-GEMM, respectively, in 5 experiments. Each histogram represents the mean of 5 independent experiments, performed in duplicates. Results are expressed as a ratio between each condition and the control condition. Vertical bars represent SD. ***P < .001. ns indicates not significant.
In contrast, exposure to 4EGI-1 barely affected the clonogenic growth or differentiation of normal CD34+ hematopoietic progenitors, even at the concentration of 100 μM. The numbers of mixed (CFU-GEMM), erythroid (BFU-E), or granulo-monocytic (CFU-GM) colonies were not significantly decreased by 4EGI-1. The broad-spectrum kinase inhibitor LY294002 was used as a control of clonogenic growth inhibition (Figure 1B).
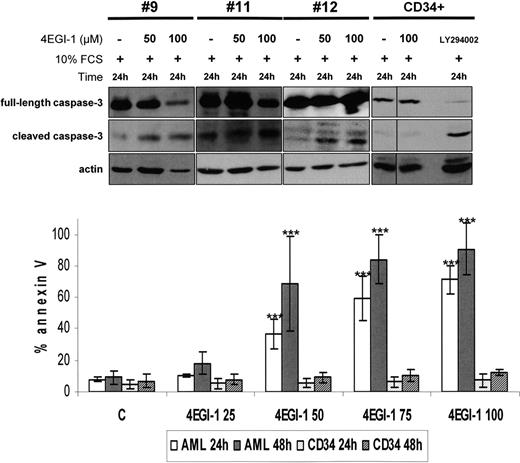
The proapoptotic potential of 4EGI-1 was further tested in 5 AML samples. 4EGI-1 was found to strongly induce apoptosis, determined by an increase of annexin-V staining and by caspase-3 cleavage (Figure 2), in a time- and dose-dependent manner in primary blast cells. Conversely, this compound had no effects on normal CD34+ hematopoietic cell survival under similar experimental conditions (Figure 2).
4EGI-1 induces a high level of apoptosis in AML blast cells but not in normal CD34+ hematopoietic progenitors. Caspase-3 cleavage: AML blast cells from 3 different AML patients were cultured 24 hours in 10% FCS MEM without or with 50 or 100 μM 4EGI-1. The same experimental procedure was applied in 3 CD34+ samples, and 25 μM LY294002 was used as a control for apoptosis induction (1 representative sample is provided). Protein extracts from 106 cells were resolved by SDS-PAGE electrophoresis, transferred to nitrocellulose, and probed with anti–caspase-3 and antiactin antibodies. Annexin V binding: 2 × 105 cells were cultured without or with increasing concentrations of 4EGI-1 (from 25 to 100μM) during 24 or 48 hours, and then labeled with annexin V–PE for flow cytometry analysis. Results are expressed as a mean of experiments done on 5 primary AML samples and on 5 different CD34+ samples. Vertical bars represent SD. ***P < .001.
4EGI-1 induces a high level of apoptosis in AML blast cells but not in normal CD34+ hematopoietic progenitors. Caspase-3 cleavage: AML blast cells from 3 different AML patients were cultured 24 hours in 10% FCS MEM without or with 50 or 100 μM 4EGI-1. The same experimental procedure was applied in 3 CD34+ samples, and 25 μM LY294002 was used as a control for apoptosis induction (1 representative sample is provided). Protein extracts from 106 cells were resolved by SDS-PAGE electrophoresis, transferred to nitrocellulose, and probed with anti–caspase-3 and antiactin antibodies. Annexin V binding: 2 × 105 cells were cultured without or with increasing concentrations of 4EGI-1 (from 25 to 100μM) during 24 or 48 hours, and then labeled with annexin V–PE for flow cytometry analysis. Results are expressed as a mean of experiments done on 5 primary AML samples and on 5 different CD34+ samples. Vertical bars represent SD. ***P < .001.
Hence, 4EGI-1 appeared to have a marked differential effect on transformed vs nontransformed hematopoietic cells, which sharply contrasted with the effects of rapamycin on primary AML cells. As previously reported by others using rapamycin,4 we confirmed that in AML, RAD001 reduced cell proliferation and CFU-L formation but did not induce apoptosis (supplemental Figure 1A-C, available on the Blood website; see the Supplemental Materials link at the top of the online article). Because translation appeared essential for AML cell survival and as mTORC1 has been shown to control the initiation step of translation through the phosphorylation of 4E-BP1 in most models, we investigated whether mTORC1 signaling could be deregulated in AML.
The phosphorylation of 4E-BP1 is independent of the activity of mTORC1 in AML blast cells
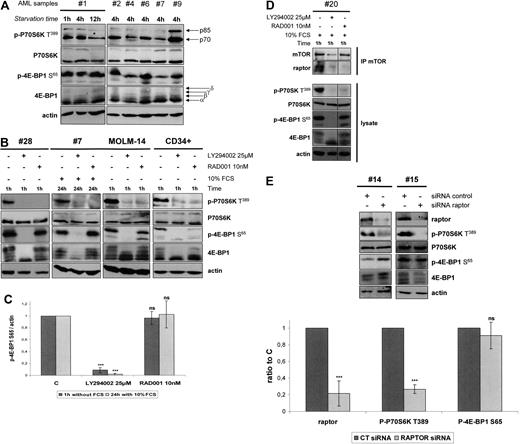
We assessed the phosphorylation status of the 2 main mTORC1 substrates, P70S6K and 4E-BP1, in 24 AML samples. As shown for sample 1, P70S6K T389 and 4E-BP1 S65 phosphorylation levels were maintained after 12 hours of serum and cytokine starvation (Figure 3A). We evaluated P70S6K and 4E-BP1 phosphorylation status after 4 hours of starvation in all samples, as reported for PI3K/Akt constitutive activity.5,29 The constitutive phosphorylation of P70S6K on T389 was detectable in 21 of 24 (87.5%) samples, and 4E-BP1 S65 phosphorylation was detected in all 24 samples tested. Western blotting analysis of 5 representative AML samples is provided in Figure 3A.
4E-BP1 phosphorylation escapes mTORC1 in AML. (A) Samples from AML patients were starved for 4 hours in cytokine- and serum-free medium, except for sample 1, starved for 1, 4, and 12 hours, and then lysed in Laemmli sampler buffer. Western blot was performed with anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti-P70S6K, anti–4E-BP1, and antiactin antibodies. Five representative samples of AML are depicted in panel A. (B) Bone marrow blast cells from 8 AML patients, cells from the MOLM-14 leukemic cell line, and normal CD34+ immature hematopoietic cells were starved for 4 hours in cytokine- and serum-free medium. A total of 10 nM RAD001 or 25 μM LY294002 was added during the last hour of starvation. Blast cells from 8 other AML patients were cultured for 24 hours in αMEM containing 10% FCS without or with 10 nM RAD001 or 25 μM LY294002. Proteins extracts from 106 cells were resolved by SDS-PAGE electrophoresis, transferred to nitrocellulose, and probed with anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti-P70S6K, anti–4E-BP1, and antiactin antibodies. (C) Western blot signals from experiments done in primary blast cells (B) were quantified using the Multi Gauge, Version 3.0 software from Fuji. Ratios of phospho-4E-BP1 S65/actin signal intensity were first calculated to correct for loading variations. Results were expressed relative to the control condition (without inhibitor) in each experiment. Each histogram represents the mean of 8 independent experiments. Vertical bars represent SD. (D) Primary AML cells were cultured without or with RAD001 during 1 hour and lysed in 0.3% CHAPS buffer. Then mTOR was immunoprecipitated (with goat anti-mTOR antibody from Santa Cruz Biotechnology), and Western blot was performed using anti-mTOR and antiraptor antibodies. Whole-cell extracts collected before mTOR immunoprecipitation were submitted to Western blots using anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti-P70S6K, anti–4E-BP1, and antiactin antibodies. (E) Bone marrow blast cells from 6 AML patients were transfected with 1 μM raptor siRNA or 1μM nontargeted control siRNA. Cell extracts were analyzed by Western blot 24 hours after electroporation to assess the expression of raptor and the phosphorylation level of P70S6K on T389 and of 4E-BP1 on S65. The intensities of phospho-4E-BP1 S65, phospho-P70S6K T389, and actin signals were quantified using the Multi Gauge, Version 3.0 software from Fuji, and ratios of phospho-4E-BP1 S65 or phospho-P70S6K T389 to actin were calculated and set to 1 for control cultures. Each histogram represents the mean of 6 independent experiments. ***P < .001. ns indicates not significant.
4E-BP1 phosphorylation escapes mTORC1 in AML. (A) Samples from AML patients were starved for 4 hours in cytokine- and serum-free medium, except for sample 1, starved for 1, 4, and 12 hours, and then lysed in Laemmli sampler buffer. Western blot was performed with anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti-P70S6K, anti–4E-BP1, and antiactin antibodies. Five representative samples of AML are depicted in panel A. (B) Bone marrow blast cells from 8 AML patients, cells from the MOLM-14 leukemic cell line, and normal CD34+ immature hematopoietic cells were starved for 4 hours in cytokine- and serum-free medium. A total of 10 nM RAD001 or 25 μM LY294002 was added during the last hour of starvation. Blast cells from 8 other AML patients were cultured for 24 hours in αMEM containing 10% FCS without or with 10 nM RAD001 or 25 μM LY294002. Proteins extracts from 106 cells were resolved by SDS-PAGE electrophoresis, transferred to nitrocellulose, and probed with anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti-P70S6K, anti–4E-BP1, and antiactin antibodies. (C) Western blot signals from experiments done in primary blast cells (B) were quantified using the Multi Gauge, Version 3.0 software from Fuji. Ratios of phospho-4E-BP1 S65/actin signal intensity were first calculated to correct for loading variations. Results were expressed relative to the control condition (without inhibitor) in each experiment. Each histogram represents the mean of 8 independent experiments. Vertical bars represent SD. (D) Primary AML cells were cultured without or with RAD001 during 1 hour and lysed in 0.3% CHAPS buffer. Then mTOR was immunoprecipitated (with goat anti-mTOR antibody from Santa Cruz Biotechnology), and Western blot was performed using anti-mTOR and antiraptor antibodies. Whole-cell extracts collected before mTOR immunoprecipitation were submitted to Western blots using anti–phospho-P70S6K T389, anti–phospho-4E-BP1 S65, anti-P70S6K, anti–4E-BP1, and antiactin antibodies. (E) Bone marrow blast cells from 6 AML patients were transfected with 1 μM raptor siRNA or 1μM nontargeted control siRNA. Cell extracts were analyzed by Western blot 24 hours after electroporation to assess the expression of raptor and the phosphorylation level of P70S6K on T389 and of 4E-BP1 on S65. The intensities of phospho-4E-BP1 S65, phospho-P70S6K T389, and actin signals were quantified using the Multi Gauge, Version 3.0 software from Fuji, and ratios of phospho-4E-BP1 S65 or phospho-P70S6K T389 to actin were calculated and set to 1 for control cultures. Each histogram represents the mean of 6 independent experiments. ***P < .001. ns indicates not significant.
We next tested the effects of RAD001 on mTORC1 signaling in AML. First, AML blast cells were cultured in starvation conditions to study their intrinsic signaling capacities. In all samples analyzed, RAD001 was found to fully inhibit the phosphorylation of P70S6K on T389 but did not decrease 4E-BP1 S65 phosphorylation. A representative Western blot is shown in Figure 3B (sample 28), and quantification of the immunoblotting signals from 8 AML samples is provided in Figure 3C. Identical results were obtained in the MOLM-14 human leukemic cell line (Figure 3B). Conversely, a complete inhibition of P70S6K T389 and 4E-BP1 S65 phosphorylation was found in normal CD34+ hematopoietic progenitors treated with RAD001 (Figure 3B, n = 3). To get closer to the activity of RAD001 in physiologic conditions, blast cells from 8 additional AML samples were cultured in 10% FCS-supplemented medium for 24 hours without or with RAD001. Under these conditions, P70S6K T389 phosphorylation was fully inhibited by RAD001, but 4E-BP1 S65 phosphorylation was unaffected. A representative Western blot (sample 7) is shown in Figure 3B. Furthermore, RAD001 inhibited neither the 4E-BP1 priming phosphorylation events on T37 and T46 nor the phosphorylation of 4E-BP1 on T70 in AML samples (supplemental Figure 2A). Finally, no inhibition of 4E-BP1 S65 phosphorylation was found when increasing the concentrations of RAD001 up to 100 nmol/L (supplemental Figure 2B). In all AML samples tested, LY294002 suppressed both P70S6K and 4E-BP1 phosphorylation.
To confirm that the disruption of the mTORC1 complex (defined by the raptor/mTOR association30 ) did not inhibit 4E-BP1 phosphorylation, we verified that RAD001 induced a dissociation of raptor from mTOR in AML cells. Indeed, we confirmed that mTOR coprecipitated with raptor in control cells but not in RAD001-treated cells, while at the same time RAD001 fully inhibited P70S6K T389 phosphorylation but did not decrease 4E-BP1 phosphorylation on S65 (Figure 3D). Lastly, we performed raptor siRNA experiments to specifically disrupt mTORC1 activity. Primary AML cells transfected with raptor siRNA showed an 80% decrease of raptor expression that paralleled the decrease of P70S6K T389 phosphorylation (P < .001). Conversely, this siRNA-induced raptor knockdown had no effect on 4E-BP1 S65 phosphorylation (Figure 3E).
These data showed that the specific blockage of mTORC1 activity in AML cells resulted in the inhibition of P70S6K phosphorylation but not of 4E-BP1 phosphorylation, thereby demonstrating that 4E-BP1 phosphorylation escaped mTORC1 control in AML.
EIF4F complex formation is abrogated in AML by 4EGI-1 but not by RAD001
Translation of mRNA is tightly regulated at the initiation level through the assembly of eIF4F complexes.18 We compared the effects of 3 potential translation inhibitors on eIF4F assembly by pull-down assays using 7m-GTP-Sepharose beads that mimic the cap structure of mRNA.8
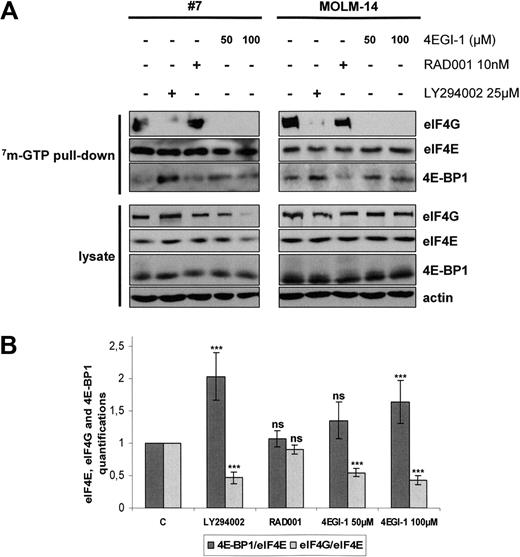
Cells were separately treated with LY294002, RAD001, and the translation inhibitor 4EGI-1. Figure 4A illustrates the results of these experiments in primary AML samples and in the MOLM-14 leukemic cell line. Quantification analysis was done for 6 independent analyses of primary AML samples (Figure 4B). Western blots using whole-cell lysates showed that these inhibitors did not significantly reduce the cellular amounts of eIF4G, eIF4E, or 4E-BP1 except for AML cells incubated with 100 μM 4EGI-1 (Figure 4A), which was probably the result of the high level of apoptosis induced by this concentration of 4EGI-1 in AML cells (Figure 2). Incubation with LY294002 increased the 4E-BP1 levels and decreased the amounts of eIF4G bound to eIF4E, indicating a major inhibition of eIF4F complex assembly and 4EGI-1 fully inhibited the interaction between eIF4E and eIF4G. Conversely, exposure to RAD001 did not modify the amounts of eIF4G or 4E-BP1 bound to eIF4E (Figure 4A-B).
EIF4F complex formation is mTORC1-independent in AML and is totally abrogated by the 4E-BP1 mimetic 4EGI-1. (A) Blast cells from 6 AML patients and from the MOLM-14 leukemic cell line were cultured for 24 hours in αMEM containing 10% FCS without or with 10 nM RAD001, 25 μM LY294002, or 50 or 100 μM 4EGI-1, and then lysed. Supernatants were clarified, m7-GTP affinity assay was performed, and beads were solubilized in boiling Laemmli sample buffer. Western blots were performed with anti-eIF4G, anti-eIF4E, and anti–4E-BP1 antibodies. (B) Quantifications of the signal intensity of Western blots from the 6 m7-GTP affinity assay experiments performed in primary AML samples were done using the Multi Gauge, Version 3.0 software from Fuji. Each histogram represents the mean of 6 independent experiments, and results are expressed as the ratio between 4E-BP1 and eIF4E and between eIF4G and eIF4E, relative to the control condition (C; extracts from cells incubated without inhibitor). Vertical bars represent SD.
EIF4F complex formation is mTORC1-independent in AML and is totally abrogated by the 4E-BP1 mimetic 4EGI-1. (A) Blast cells from 6 AML patients and from the MOLM-14 leukemic cell line were cultured for 24 hours in αMEM containing 10% FCS without or with 10 nM RAD001, 25 μM LY294002, or 50 or 100 μM 4EGI-1, and then lysed. Supernatants were clarified, m7-GTP affinity assay was performed, and beads were solubilized in boiling Laemmli sample buffer. Western blots were performed with anti-eIF4G, anti-eIF4E, and anti–4E-BP1 antibodies. (B) Quantifications of the signal intensity of Western blots from the 6 m7-GTP affinity assay experiments performed in primary AML samples were done using the Multi Gauge, Version 3.0 software from Fuji. Each histogram represents the mean of 6 independent experiments, and results are expressed as the ratio between 4E-BP1 and eIF4E and between eIF4G and eIF4E, relative to the control condition (C; extracts from cells incubated without inhibitor). Vertical bars represent SD.
We conclude from these analyses that mTORC1 inhibition by RAD001 failed to inhibit eIF4F assembly.
4EGI-1 but not RAD001 reduces the translation of c-Myc mRNA in the MOLM-14 AML cell line
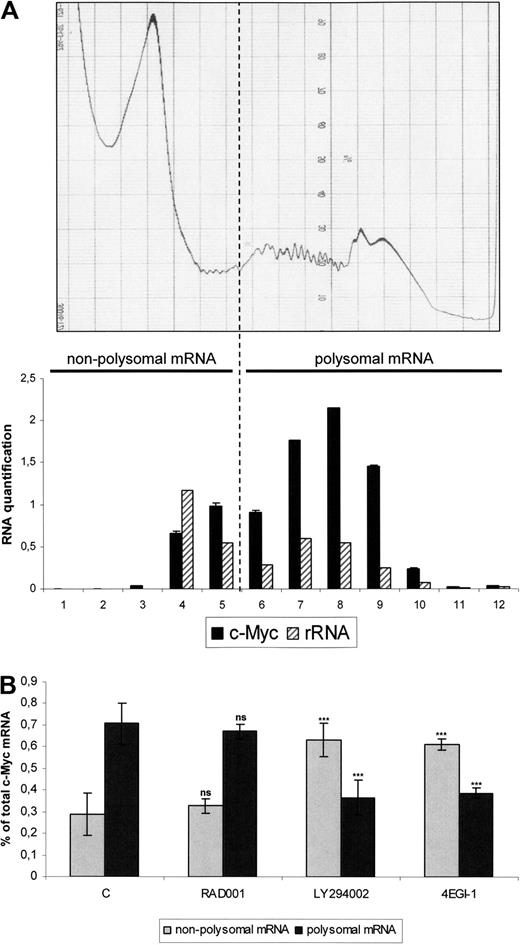
C-Myc mRNA is a “weak mRNA,” characterized by a long structured 5′UTR. Its efficient translation requires a high level of active translation initiation complexes.24 We determined the levels of c-Myc associated with polysomes, therefore engaged in translation. MOLM-14 cells were treated with LY294002, RAD001, or 4EGI-1, and extracts were separated by centrifugation through sucrose gradients. We determined that free ribosomes had sedimented in fractions 4 and 5 and polysomes in fractions 6 to 12 by coupling the results of continuous ultraviolet (UV) absorbance measurement and ribosomal RNA quantifications in the 12 fractions separately (Figure 5A). The results of 3 independent experiments of c-Myc quantification are presented in Figure 5B. In the control cells, 76% of the c-Myc mRNA molecules were found to be associated with polysomes and thus undergoing active translation. The proportion of these transcripts in the polysome-bound fraction was not significantly decreased by RAD001. In contrast, most mRNAs were present in the free fraction in LY294002- or 4EGI-1-treated cells (Figure 5B), and this effect was more pronounced for c-Myc mRNA than for actin (data not shown). Therefore, these results show that mTORC1 inhibition by RAD001 did not decrease the translation efficiency of oncogenic mRNAs, such as c-Myc in leukemic cells, in contrast to LY294002 or 4EGI-1.
4EGI-1 but not RAD001 reduces the translation rate of c-Myc mRNA in the MOLM-14 AML cell line. Cells from the MOLM-14 leukemic cell line were cultured 12 hours at 106/mL in αMEM containing 10% FCS, without or with 10 nM RAD001, 25 μM LY294002, or 100 μM 4EGI-1, and then cell lysates from 108 cells were subjected to sucrose density centrifugation and processed as described in “Polysome analysis.” Gradients were fractioned using a Beckman fraction recovery system connected to a LKB UV detector. UV recording is shown in the top level of the figure. Twelve fractions were collected, and ribosome content of each fraction was determined by Agilent 2100 electrophoresis for both 18S and 28S rRNA (A, bottom panel). RNA was extracted in each fraction separately and quantitative reverse-transcribed PCR for c-Myc expression was performed; results are expressed together with rRNA quantifications results in the lower panel of the panel A. From these experiments, we determined that free ribosome subunits sedimented in fractions 4 and 5. Accordingly, fractions 6 to 12 were considered to contain polysomes. The nonpolysome (fractions 1-5– and polysome (fractions 6-12)–containing fractions were then pooled and analyzed by real-time quantitative PCR for c-Myc mRNAs. Results are expressed as a percentage of nonpolysomal-bound or polysomal-bound c-Myc mRNA relative to the total c-Myc mRNA amount (B).
4EGI-1 but not RAD001 reduces the translation rate of c-Myc mRNA in the MOLM-14 AML cell line. Cells from the MOLM-14 leukemic cell line were cultured 12 hours at 106/mL in αMEM containing 10% FCS, without or with 10 nM RAD001, 25 μM LY294002, or 100 μM 4EGI-1, and then cell lysates from 108 cells were subjected to sucrose density centrifugation and processed as described in “Polysome analysis.” Gradients were fractioned using a Beckman fraction recovery system connected to a LKB UV detector. UV recording is shown in the top level of the figure. Twelve fractions were collected, and ribosome content of each fraction was determined by Agilent 2100 electrophoresis for both 18S and 28S rRNA (A, bottom panel). RNA was extracted in each fraction separately and quantitative reverse-transcribed PCR for c-Myc expression was performed; results are expressed together with rRNA quantifications results in the lower panel of the panel A. From these experiments, we determined that free ribosome subunits sedimented in fractions 4 and 5. Accordingly, fractions 6 to 12 were considered to contain polysomes. The nonpolysome (fractions 1-5– and polysome (fractions 6-12)–containing fractions were then pooled and analyzed by real-time quantitative PCR for c-Myc mRNAs. Results are expressed as a percentage of nonpolysomal-bound or polysomal-bound c-Myc mRNA relative to the total c-Myc mRNA amount (B).
Oncogenic protein accumulation is abrogated by 4EGI-1 but not by RAD001 in AML cells
To further study the impact of RAD001, LY294002, and 4EGI-1 on translation at the protein level, we analyzed the global protein synthesis in 5 AML samples using a [3H]leucine pulse assay. Whereas RAD001 did not significantly decrease global protein synthesis, LY294002 led to a 42% decrease in [3H]leucine incorporation (P < .001) and 4EGI-1 induced a major decrease of [3H]leucine incorporation in primary AML samples (decrease of 78%, P < .001, Figure 6A). Moreover, the expression of proteins widely implicated in oncogenic processes and regulated at the translation initiation level, c-Myc, Cyclin D1, and Bcl-xL, was examined by Western blotting in 5 AML samples cultured during 24 hours in 10% FCS media without or with 10 nmol/L RAD001, or 50 or 100 μM 4EGI-1. In all samples treated with RAD001, the amounts of these proteins were identical to the control condition. Conversely, 4EGI-1 dramatically reduced the accumulation of these oncogenic proteins (from 60%-80%, P < .001). Analysis of a representative sample (sample 2) and data quantification from 5 independent experiments are shown in Figure 6B. Therefore, we confirmed that 4EGI-1 but not RAD001 induced a major decrease in the translation of malignancy-related mRNAs in primary AML cells.
Translation is reduced by 4EGI-1 but not by RAD001 in AML. (A) [3H] leucine pulses were performed to determine global protein synthesis rates. Blast cells from 5 AML patients and from the MOLM-14 leukemic cell line were cultured for 2 hours in a leucine-deficient medium, without or with 10 nM RAD001, 25 μM LY294002, or 50 or 100 μM 4EGI-1, and then pulsed with [3H] leucine (1 μCi, 37 kBq) for 1 hour. The amount of radioactivity incorporated into macromolecules was determined by trichloroacetic acid precipitation. Results are presented as the ratio to the control incubation without inhibitor. Vertical bars represent SD. (B) Blast cells from 5 AML patients were cultured for 24 hours in αMEM containing 10% FCS, without or with 10 nM RAD001 or 50 or 100 μM 4EGI-1. Cell extracts were analyzed by Western blot to assess the expression of c-Myc, Cyclin D1, Bcl-xL, and actin. The signal intensity was quantified, and results are expressed as the ratio between c-Myc, Cyclin D1, or Bcl-xL and actin signals, relative to the control condition. Vertical bars represent SD.
Translation is reduced by 4EGI-1 but not by RAD001 in AML. (A) [3H] leucine pulses were performed to determine global protein synthesis rates. Blast cells from 5 AML patients and from the MOLM-14 leukemic cell line were cultured for 2 hours in a leucine-deficient medium, without or with 10 nM RAD001, 25 μM LY294002, or 50 or 100 μM 4EGI-1, and then pulsed with [3H] leucine (1 μCi, 37 kBq) for 1 hour. The amount of radioactivity incorporated into macromolecules was determined by trichloroacetic acid precipitation. Results are presented as the ratio to the control incubation without inhibitor. Vertical bars represent SD. (B) Blast cells from 5 AML patients were cultured for 24 hours in αMEM containing 10% FCS, without or with 10 nM RAD001 or 50 or 100 μM 4EGI-1. Cell extracts were analyzed by Western blot to assess the expression of c-Myc, Cyclin D1, Bcl-xL, and actin. The signal intensity was quantified, and results are expressed as the ratio between c-Myc, Cyclin D1, or Bcl-xL and actin signals, relative to the control condition. Vertical bars represent SD.
Altogether, our results show that the control of protein synthesis escaped mTORC1 control in AML. Nevertheless, results of experiments using LY294002 suggested that another kinase may regulate 4E-BP1 phosphorylation and protein synthesis. LY294002 is a broad-spectrum inhibitor of class 1A PI3K. However, specifically blocking PI3K activity in AML cells using the p110δ inhibitor IC87114 did not inhibit 4E-BP1 phosphorylation, even in the presence of rapamycin (supplemental Figure 3). LY294002 has also been reported to inhibit the activity of Pim kinases.31 Interestingly, the Pim-2 kinase has been implicated in the control of 4E-BP1 phosphorylation.32,33 We therefore tested whether this kinase could be involved in 4E-BP1 phosphorylation and in the control of protein synthesis in AML cells.
The Pim-2 serine/threonine kinase is constitutively expressed in AML blast cells and controls 4E-BP1 S65 phosphorylation and c-Myc and Cyclin D1 expression independently of mTORC1
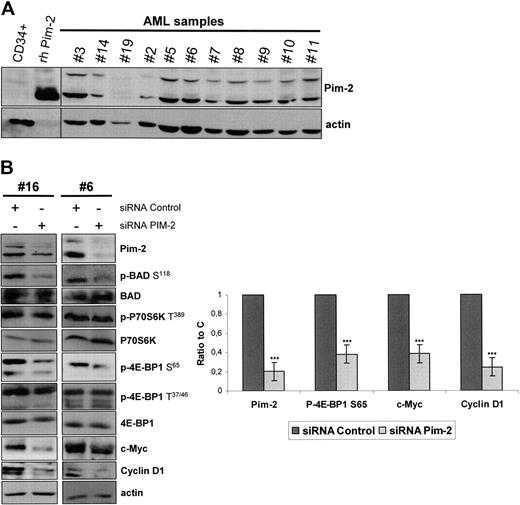
We tested the expression of Pim-2, for which isoforms of 34, 37, and 40 kDa have been reported,32,34 in 24 highly infiltrated marrow samples from AML patients. Pim-2 was found to be expressed in 23 of 24 samples tested (96%), with a variable participation of the 34- and 40-kDa isoforms, whereas the expression of the 37-kDa isoform appeared weak. Eleven representative samples are presented in Figure 7A. In contrast, Pim-2 was barely detectable in normal CD34+ hematopoietic progenitors (Figure 7A).
Pim-2 is constantly expressed in AML and controls 4E-BP1 S65 phosphorylation and subsequent cap-dependent translation in AML. (A) Samples from AML patients or normal CD34+ hematopoietic cells from healthy donors were starved for 4 hours in cytokine- and serum-free medium. A total of 106 cells were lysed in boiling Laemmli sample buffer and Western blot for Pim-2 was performed, with 50 ng of human recombinant Pim-2 (rh Pim-2, from Cell Signaling Technology) as control for Pim-2 detection. Actin was used as the loading control. (B) Pim-2 siRNA or nontargeted control siRNA were transfected into AML blast cells, with the Amaxa nucleofector system. At 24 hours after electroporation, cell extracts were analyzed by Western blot to assess the expression of Pim-2, c-Myc, Cyclin D1, P70S6K, 4E-BP1, BAD, and actin and the phosphorylation level of P70S6K on T389, BAD on S118, and of 4E-BP1 on T37/46 and S65 residues. The signal intensity was quantified using the Multi Gauge, Version 3.0 software from Fuji. Each histogram represents the mean of 4 independent experiments, and results are expressed as a ratio between the Pim-2, c-Myc, Cyclin D1, or the phospho-4E-BP1 S65 signals and the actin signal, relative to the control condition. Vertical bars represent SD.
Pim-2 is constantly expressed in AML and controls 4E-BP1 S65 phosphorylation and subsequent cap-dependent translation in AML. (A) Samples from AML patients or normal CD34+ hematopoietic cells from healthy donors were starved for 4 hours in cytokine- and serum-free medium. A total of 106 cells were lysed in boiling Laemmli sample buffer and Western blot for Pim-2 was performed, with 50 ng of human recombinant Pim-2 (rh Pim-2, from Cell Signaling Technology) as control for Pim-2 detection. Actin was used as the loading control. (B) Pim-2 siRNA or nontargeted control siRNA were transfected into AML blast cells, with the Amaxa nucleofector system. At 24 hours after electroporation, cell extracts were analyzed by Western blot to assess the expression of Pim-2, c-Myc, Cyclin D1, P70S6K, 4E-BP1, BAD, and actin and the phosphorylation level of P70S6K on T389, BAD on S118, and of 4E-BP1 on T37/46 and S65 residues. The signal intensity was quantified using the Multi Gauge, Version 3.0 software from Fuji. Each histogram represents the mean of 4 independent experiments, and results are expressed as a ratio between the Pim-2, c-Myc, Cyclin D1, or the phospho-4E-BP1 S65 signals and the actin signal, relative to the control condition. Vertical bars represent SD.
As no specific cell-permeable Pim-2 inhibitor is available yet, we achieved Pim-2 knockdown in 6 AML samples by siRNA, and a clear decrease of 4E-BP1 S65 phosphorylation paralleled the reduction of Pim-2 expression in the transfected cells (Figure 7B). Interestingly, Pim-2 knockdown also inhibited the phosphorylation of the proapoptotic protein BAD on S118 (Figure 7B). Indeed, BAD S118 (equivalent to S112 in murine BAD) has been previously described as a direct substrate for Pim-2.35 Interestingly, P70S6K T389 phosphorylation was unaffected by Pim-2 knockdown (Figure 7B), demonstrating that Pim-2 stimulates 4E-BP1 phosphorylation on S65 independently of mTORC1. Lastly, we observed that Pim-2 knockdown did not reduce the priming phosphorylation of 4E-BP1 on T37/46 (Figure 7B).
As 4E-BP1 S65 phosphorylation is absolutely required for cap-dependent translation initiation,23 we assumed that Pim-2 could control protein synthesis in AML cells. To strengthen this hypothesis, we tested the effects of Pim-2 knockdown on the accumulation of oncogenic proteins regulated at the translation initiation level. We indeed observed that Pim-2 siRNA markedly reduced c-Myc and Cyclin D1 accumulation (Figure 7B, n = 4) without affecting the amounts of the corresponding mRNAs (data not shown).
Discussion
A prevailing model suggests that cancers that are dependent on the activation of the Akt oncoprotein rely on the subsequent activation of mTORC1 to drive tumorigenesis, leading to the clinical development of rapamycin as a cancer drug.36 However, in AML, despite a frequent activation of mTORC1, rapamycin is essentially cytostatic and only enhances chemotherapy-induced apoptosis,4 which markedly contrasts with the expected effects of mTORC1 inhibition because this complex is thought to control cap-dependent mRNA translation, which is essential for cancer cells.
In this report, we established that the control of translation is of major importance for AML biology using the 4EGI-1 compound, which pharmacologically mimics the 4E-BP1 translation repressor activity.27 When used for the first time in primary AML samples, 4EGI-1 markedly impaired the clonogenic growth of leukemic progenitors and massively induced apoptosis of blast cells; interestingly, these effects occurred with a minimal degree of toxicity against normal hematopoietic processes.
Because the concentrations of 4EGI-1 required to inhibit protein synthesis and to induce apoptosis are rather high (Moerke et al27 and our present results), we sought for off-target effects of this compound. As shown in supplemental Figure 3, 4EGI-1 neither decreased Akt, Erk, and mTORC1 activity nor induced caspase-3 cleavage during the first 8 hours of cell incubation, although protein synthesis was strongly inhibited from 2 hours of exposure to 4EGI-1 (Figure 6A), suggesting that apoptosis observed in AML cells after 24 hours of incubation is the direct consequence of translation inhibition rather than off-target effects of 4EGI-1.
As the antileukemic activity of 4EGI-1 was dramatically superior to the effects of RAD001 in AML,4,13 we studied mTORC1 signaling in primary AML samples. We examined 4E-BP1 phosphorylation and particularly focused on its S65 residue, which is phosphorylated in last and is absolutely required to abrogate the repressor activity of 4E-BP1 against eIF4F formation.23 In most models, 4E-BP1 S65 phosphorylation is strongly regulated by growth factors and is reported as a rapamycin-sensitive event.23,37 We demonstrated herein that the phosphorylation of 4E-BP1 was not decreased by RAD001 or raptor siRNA in AML. Because the mTORC1 definition is based on the raptor/mTOR interaction,30,38 our interpretation is that 4E-BP1 phosphorylation on S65 escapes mTORC1 control in AML cells, and indeed we show here that 4E-BP1 S65 phosphorylation is dependent on Pim-2 expression. These results are in discrepancy with previous reports showing in a small number of AML samples that treatment with rapalogs may reduce 4E-BP1 T70 phosphorylation in AML cells.5,13 This could be explained in part by the very high concentrations of rapalogs generally used in these works.4,30
Our result that rapalogs did not control 4E-BP1 S65 phosphorylation in AML cells is supported by a recent work by Choo et al showing a recovery of 4E-BP1 phosphorylation but not of P70S6K phosphorylation in 293T cells cultured with rapamycin for at least 6 hours.39 Another recent work by Thoreen et al shows that, in MEF cells, 4E-BP1 phosphorylation is also rapamycin-resistant.40 However, in both cases, 4E-BP1 phosphorylation requires raptor and not Pim-2, which is different from the mechanisms we observed in AML cells. Nevertheless, the regulation of 4E-BP1 phosphorylation is complex, and we do not rule out that a raptor-independent activity of mTOR may be required for 4E-BP1 T37/46 priming phosphorylation events.21-23 Interestingly, we observed that these priming events were abolished by PI-103, a dual PI3K and mTOR catalytic inhibitor,13,15 and by mTOR siRNA (data not shown). This suggests that mTOR, in a rapamycin-insensitive and raptor-independent complex, could be responsible for the priming phosphorylation of 4E-BP1.
We further demonstrated the absence of control of mRNA translation by mTORC1 by comparing the effects of RAD001 to the specific translation inhibitor 4EGI-1. 4EGI-1, in contrast to RAD001, induced a complete inhibition of the association between eIF4G and eIF4E in AML cells. Therefore, unlike RAD001, 4EGI-1: (1) reduced the levels of c-Myc mRNA bound to polysomes, (2) decreased overall protein synthesis, and (3) decreased the accumulation of oncogenic proteins regulated at the translation initiation level (eg, c-Myc, Cyclin D1, and Bcl-xL). We confirmed that the decrease of oncogenic proteins expression we observed was the result of a specific blockade of mRNA translation as the amount of c-Myc mRNA was not modified in 4EGI-1 treated cells (data not shown). Furthermore, the decrease of c-Myc and Cyclin D1 expression did not result of a caspase-mediated proteolytic cleavage as the decrease in the expression of these proteins was not reverted by the caspase inhibitor Z-VAD in the MOLM-14 leukemic cell line (supplemental Figure 5). Our data suggest that the modest activity of rapamycin previously observed in vitro and in vivo in AML4,13 might be explained in part by the loss of mTORC1 control on oncoprotein synthesis.
Our results indicate the Pim-2 serine/threonine kinase as an essential regulator of 4E-BP1 phosphorylation in AML. The Pim kinase family includes 3 serine/threonine kinases (Pim1, 2, and 3) strongly involved in oncogenesis and whose activity appears to be constitutive. The Pim1 and Pim2 genes were first identified as frequent MoMuLV proviral insertion sites in murine lymphomas.41,42 Moreover, overexpression of Pim1 or Pim2 in transgenic mice results in a high rate of lymphomas,43,44 and these kinases are required for pre-B transformation by v-Abl.45 Pim-2 is thought to be involved in the growth and survival of nontransformed hematopoietic cells independently of the PI3K/Akt pathway, and primary hematopoietic cells from Pim2−/− mice are hypersensitive to rapamycin.33 Interestingly, Pim-2 complements wtFLT3 to transform 32D cells, suggesting that it is an essential effector of myeloid leukemogenesis.46,47 Pim kinases have also been implicated in AML cells survival and targeted therapy suppresses in vitro growth of leukemic cells.48
When tested in 24 AML samples, Pim-2 was detected in 23 cases, whereas its expression was markedly low in normal CD34+ cells. Interestingly, the Pim-1 protein was seldom found expressed in AML cells (data not shown), which illustrated the nonredundant functions of these kinases.33 In primary AML cells, the siRNA-mediated decrease of Pim-2 expression paralleled the decrease of 4E-BP1 S65 phosphorylation, but P70S6K T389 phosphorylation levels remained unchanged, demonstrating that Pim-2 controlled 4E-BP1 phosphorylation in an mTORC1-independent way. Using an in vitro kinase assay, we did not detect any direct phosphorylation of 4E-BP1 by Pim-2, suggesting that other(s) yet unidentified kinase(s) activated downstream Pim-2 are involved in this process (data not shown). This hypothesis is enforced by analysis of the sequence surrounding 4E-BP1 S65, which does not show homology with the consensus sequence determined for Pim-2 phosphorylation (RXRHXS).49 Interestingly, we observed that Pim-2 regulates the synthesis of proteins encoded by oncogene-related mRNAs in AML as Pim-2 siRNA decreases 4E-BP1 S65 phosphorylation in parallel with a suppression of oncoproteins controlled at the translation initiation level (c-Myc and Cyclin D1).
Targeting translation demonstrated a favorable therapeutic index ex vivo in AML. Indeed, even if normal CD34+ hematopoietic progenitors are efficiently targeted by 4EGI-1 (data not shown), this compound did not induce significant apoptosis of these cells. Furthermore, the clonogenic growth and differentiation of CD34+ cells were preserved, suggesting that very immature progenitor cells among the normal CD34+ compartment were not affected by 4EGI-1. Graff et al have also recently reported that cancer tissues are more susceptible than normal tissues to translation inhibition using anti-eIF4E antisense oligonucleotides.50
In conclusion, our results demonstrate that cap-dependent translation leading to the synthesis of highly oncogenic proteins is deregulated in AML. This process escapes mTORC1 and is controlled in part by a Pim-2/4E-BP1 axis, which explains both the absence of translation inhibition by rapamycin and its weak clinical efficacy in AML (supplemental Figure 6, schematic overview). We strongly suggest that direct targeting of the translation initiating complex using 4EGI-1 or other approaches, such as anti-eIF4E antisense oligonucleotides50 or specific Pim-2 inhibitors when available, should have a significant impact on the development of novel AML therapies.
An Inside Blood analysis of the article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all participating investigators from the GOELAMS as well as Dr G. Wagner (Harvard Medical School, Boston, MA) who kindly provided 4EGI-1, and Drs Y. Zermati (Institut Cochin, Paris, France), O. Kosmider (Laboratoire d'hématologie, Hôpital Cochin, Paris, France), and F. Verdier (Institut Cochin, Paris, France) for helpful criticism of the manuscript
This work was supported by grants from the Ligue Nationale Contre le Cancer (laboratoire associé), the Institut National du Cancer, and the Association Laurette Fugain, Paris, France. J.T. and A.S.G. were supported by the Fondation pour la Recherche Medicale, Paris, France. S.P. was supported by AP-HP/La Caisse Nationale d'Assurance Maladie, Paris, France. N.C. was supported by Inserm.
A complete list of GOELAMS participants appears in the supplemental Appendix.
Authorship
Contribution: J.T. designed and performed research, analyzed data, and wrote the manuscript; A.S.G, V.B., N.C., S.P., L.W., and M.U. performed research and analyzed data; N.I. and F.D. contributed AML patient samples and analyzed clinical data; C.L. and P.M. analyzed data and wrote the manuscript; and D.B. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Didier Bouscary, Institut Cochin, Inserm U567, 27 rue du Faubourg Saint Jacques, 75014 Paris, France; e-mail: didier.bouscary@inserm.fr.

![Figure 6. Translation is reduced by 4EGI-1 but not by RAD001 in AML. (A) [3H] leucine pulses were performed to determine global protein synthesis rates. Blast cells from 5 AML patients and from the MOLM-14 leukemic cell line were cultured for 2 hours in a leucine-deficient medium, without or with 10 nM RAD001, 25 μM LY294002, or 50 or 100 μM 4EGI-1, and then pulsed with [3H] leucine (1 μCi, 37 kBq) for 1 hour. The amount of radioactivity incorporated into macromolecules was determined by trichloroacetic acid precipitation. Results are presented as the ratio to the control incubation without inhibitor. Vertical bars represent SD. (B) Blast cells from 5 AML patients were cultured for 24 hours in αMEM containing 10% FCS, without or with 10 nM RAD001 or 50 or 100 μM 4EGI-1. Cell extracts were analyzed by Western blot to assess the expression of c-Myc, Cyclin D1, Bcl-xL, and actin. The signal intensity was quantified, and results are expressed as the ratio between c-Myc, Cyclin D1, or Bcl-xL and actin signals, relative to the control condition. Vertical bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/8/10.1182_blood-2008-10-184515/4/m_zh89990939250006.jpeg?Expires=1768509854&Signature=uH6KrD4-5~r7ZYxr36KHrXb3MSauFZsRSrOfZg4X8DLhO6BFTHd0xffrCPuQy5bS1RB2aTGVc6GgDMINrBpUqt1XYd1dkKh5GpDjEnzxDWVwBOpX2HqdThofsH3PyTE808-nVej5afOgbqTnC1O9o0iBDvTfizw1XpjkR33K8MFE~uCSZ-O3vHAS8YCpV-yL98HIz7GLP6MKRxbZu9PLxSuGN6B1vgbuQap1lnfPHZ295KdiKMClEO3glqToTNalx3N92ltsnevdtej5HqgatAt2DBivPuC9Roz9FV7gUrAhwhTijMHYFiptThI9-~JQH9tI3Vr6Wy8w~UcqTPdrAg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)