Abstract
T-cell acute lymphoblastic leukemia 1 (TAL1), also known as stem cell leukemia (SCL), plays important roles in differentiation of hematopoietic and endothelial cells and is deregulated in a high percentage of T-cell acute lymphoblastic leukemia (T-ALL). In this report we show that the intracellular concentration of TAL1 is regulated by transforming growth factor β (TGF-β), which triggers its polyubiquitylation and degradation by the proteasome. This effect is mediated by AKT1, which phosphorylates TAL1 at threonine 90. Immunoprecipitation experiments showed that this event increases association of TAL1 with the E3 ubiquitin ligase CHIP. The E47 heterodimerization partner of TAL1 hinders this association. Our observations indicate that activation of the TGF-β and phosphatidylinositol 3-kinase/AKT pathways might reverse overexpression of TAL1 in leukemic cells by inducing proteolysis of this important oncogene.
Introduction
T-cell acute lymphoblastic leukemia 1 (TAL1) is normally expressed in early hematopoiesis and is essential for generation of the erythroid and myeloid lineages.1 In T-cell lineage TAL1 expression is normally repressed but is deregulated in a high percentage of T-cell acute lymphoblastic leukemia (T-ALL) as a consequence of either chromosomal rearrangement placing the TAL1 gene under SIL promoter control2,3 or epigenetic activation.4 Like most tissue-specific basic helix-loop-helix (bHLH) factors, TAL1 binds to E-box motifs as a heterodimer with E proteins such as E47, and it can nucleate various activatory or inhibitory protein complexes.5-9 RNA interference experiments have established that silencing of TAL1 in the Jurkat T-ALL cell line impairs their proliferation.8 Hence, a possible therapeutic approach would be to cause removal of this oncogene in T-ALL cells abnormally expressing it. Toward this end, we have examined factors controlling stability of this protein. In this report we show that transforming growth factor β (TGF-β) causes polyubiquitylation and proteasome-mediated degradation of TAL1. This effect relies on TAL1 phosphorylation by AKT1, an event that increases association of the oncogene with the E3 ubiquitin ligase CHIP. This TGF-β–induced modification of TAL1 is inhibited by heterodimerization with its partner E47.
Methods
Cell culture and transfection
Cell culture and transfection conditions were previously described.10 To achieve inhibition of the proteasome, MG132 (Sigma-Aldrich, St Louis, MO) was added at 10 μM for 6 hours. Phosphatidylinositol 3-kinase (PI3K)–specific inhibitor wortmannin was dissolved in dimethyl sulfoxide (DMSO) and added in the medium at 10 μM 30 minutes before treatment with human platelet TGF-β1 (1-10 ng/mL) for 9 hours. Cellular extracts were normalized with respect to protein concentrations, which were quantified with the DC protein assay kit (Bio-Rad, Hercules, CA).
Immunoblot
Primary antibodies were diluted 1:1000 or as indicated by the manufacturer, and detection was performed by chemiluminescence using enhanced chemiluminescence (ECL) or ECL Plus kits (GE Healthcare, Little Chalfont, United Kingdom). Monoclonal antibody against TAL1 (3BTL73) was provided by D. Mathieu.11 The following antibodies were purchased: FLAG (clone M2; Sigma-Aldrich), β-actin (Sigma-Aldrich), Myc (clone 9E10; Sigma-Aldrich), and hemagglutinin (HA, clone 12CA5; Roche, Basel, Switzerland).
Real-time quantitative reverse transcription–polymerase chain reaction
Total RNAs were extracted from frozen cells using the RNeasy Mini kit (QIAGEN, Valencia, CA). One-step reverse transcription–polymerase chain reaction (RT-PCR) reactions were performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen) and the LightCycler apparatus (Roche). Sequences of sense and antisense primers were described previously.10
Results and discussion
TGF-β induces degradation of TAL1
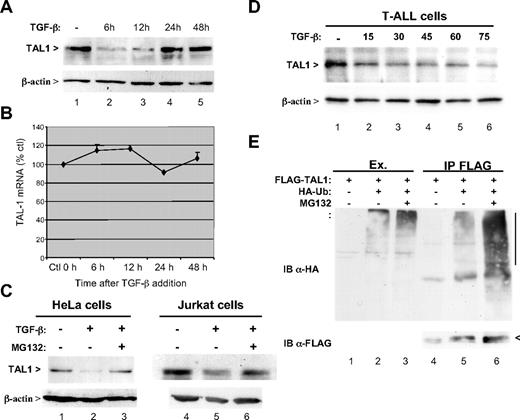
After TGF-β addition, Jurkat cells protein extracts were prepared at various times and analyzed by immunoblot using a monoclonal antibody to TAL1. At 6 hours, a marked reduction in the amount of intracellular TAL1 was apparent (Figure 1A top panel lane 2). It persisted at 12 hours and then disappeared at 24 and 48 hours, likely as a consequence of receptor internalization or induction of inhibitory Smad7. Under these conditions no modification of β-actin concentration taken as a control was observed (Figure 1A bottom panel). It was then investigated whether this decrease was related to TAL1 mRNA amount. Quantification of this mRNA did not show any significant variation (Figure 1B), ruling out this possibility. To test whether this TGF-β–dependent decrease in TAL1 might be due to induced protein degradation, the effect of the MG132 proteasome inhibitor was examined. Both TAL1 expressed in HeLa cells and endogenous TAL1 in Jurkat cells were protected from TGF-β–induced inhibition when treated with MG132 (Figure 1C top panel). It was also tested whether this effect occurs in cells taken from T-ALL patients. As shown in Figure 1D, a dose-dependent reduction in TAL1 intracellular concentration was also observed using cells from a patient with a SIL-TAL1 rearrangement. These observations support the notion that TGF-β strongly reduces the intracellular amount of TAL1 by inducing its degradation by the proteasome.
TGF-β induces the proteasome-dependent degradation of TAL1. (A) Decreased expression of TAL1 in the presence of TGF-β. Jurkat cells were treated with 10 ng/mL TGF-β during indicated times, and extracts were prepared from these cells were analyzed by immunoblot using antibodies to TAL1 (top panel) or to β-actin, used here as loading control (bottom panel). (B) TAL1 transcription is not affected by TGF-β. Total RNAs from TGF-β–treated Jurkat cells were purified and analyzed by real-time quantitative RT-PCR for TAL1 mRNA. Quantification was performed with respect to untreated cells and normalized by β-actin mRNA. Mean of 3 measures is represented with standard deviation as percentage with respect to untreated cells. (C) MG132 reverses TGF-β-induced degradation of TAL1. HeLa cells transfected with pSGF-TAL110 (lanes 1 to 3) or Jurkat cells (lanes 4-6) were treated with 10 ng/mL of TGF-β during 9 hours and with or without addition of 10 μM MG132 during the last 6 hours. Cellular extracts were normalized with respect to protein concentration and analyzed by immunoblot using antibodies to TAL1 (top panel) or to β-actin (bottom panel). (D) TGF-β-induced degradation of TAL1 in leukemic cells. Either fresh or frozen cells from 5 patients suffering T-ALL with TAL1 deregulation (4 SIL-TAL1 rearrangement and one translocation t(1;14)) were cultured in RPMI 1640 plus 10% fetal calf serum and were treated without or with 15, 30, 45, 60, and 75 ng/mL TGF-β for 6 hours. Extracts were prepared in radioimmunoprecipitation assay buffer and equal total protein amounts were analyzed by immunoblot as described for panel C. Results from one representative patient are shown. (E) TAL1 is polyubiquitylated. HeLa cells were transfected with 1 μg of pSGF-TAL1 and 0.5 μg of pSG-HA-Ub,12 with or without MG132 treatment as indicated. Cell lysates were immunoprecipitated with an antibody to FLAG. Immunoblot analysis of extracts (lanes 1-3) was done using the antibody to HA and that of immunoprecipitates (lanes 4-6) using antibodies to HA (top panel) and FLAG (bottom panel).
TGF-β induces the proteasome-dependent degradation of TAL1. (A) Decreased expression of TAL1 in the presence of TGF-β. Jurkat cells were treated with 10 ng/mL TGF-β during indicated times, and extracts were prepared from these cells were analyzed by immunoblot using antibodies to TAL1 (top panel) or to β-actin, used here as loading control (bottom panel). (B) TAL1 transcription is not affected by TGF-β. Total RNAs from TGF-β–treated Jurkat cells were purified and analyzed by real-time quantitative RT-PCR for TAL1 mRNA. Quantification was performed with respect to untreated cells and normalized by β-actin mRNA. Mean of 3 measures is represented with standard deviation as percentage with respect to untreated cells. (C) MG132 reverses TGF-β-induced degradation of TAL1. HeLa cells transfected with pSGF-TAL110 (lanes 1 to 3) or Jurkat cells (lanes 4-6) were treated with 10 ng/mL of TGF-β during 9 hours and with or without addition of 10 μM MG132 during the last 6 hours. Cellular extracts were normalized with respect to protein concentration and analyzed by immunoblot using antibodies to TAL1 (top panel) or to β-actin (bottom panel). (D) TGF-β-induced degradation of TAL1 in leukemic cells. Either fresh or frozen cells from 5 patients suffering T-ALL with TAL1 deregulation (4 SIL-TAL1 rearrangement and one translocation t(1;14)) were cultured in RPMI 1640 plus 10% fetal calf serum and were treated without or with 15, 30, 45, 60, and 75 ng/mL TGF-β for 6 hours. Extracts were prepared in radioimmunoprecipitation assay buffer and equal total protein amounts were analyzed by immunoblot as described for panel C. Results from one representative patient are shown. (E) TAL1 is polyubiquitylated. HeLa cells were transfected with 1 μg of pSGF-TAL1 and 0.5 μg of pSG-HA-Ub,12 with or without MG132 treatment as indicated. Cell lysates were immunoprecipitated with an antibody to FLAG. Immunoblot analysis of extracts (lanes 1-3) was done using the antibody to HA and that of immunoprecipitates (lanes 4-6) using antibodies to HA (top panel) and FLAG (bottom panel).
TAL1 modification by polyubiquitylation
We further examined whether TAL1 was modified by polyubiquitylation. HeLa cells were transfected with vectors expressing FLAG-tagged TAL1 and HA-tagged ubiquitin. TAL1 was immunoprecipitated and analyzed by immunoblot using an antibody to HA to detect ubiquitin chains associated with TAL1. This analysis indeed revealed a smear of bands above the normal position of TAL1 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), which was strongly increased by treatment of the cells with the proteasome inhibitor MG132 (Figure 1E). Furthermore, it was observed that TGF-β treatment is able to increase the endogenous polyubiquitylation of TAL1 (J.-M. T., unpublished data, September 2008). These results indicated that degradation of TAL1 induced by TGF-β correlates with its polyubiquitylation.
TGF-β–induced degradation of TAL1 involves its phosphorylation by AKT1
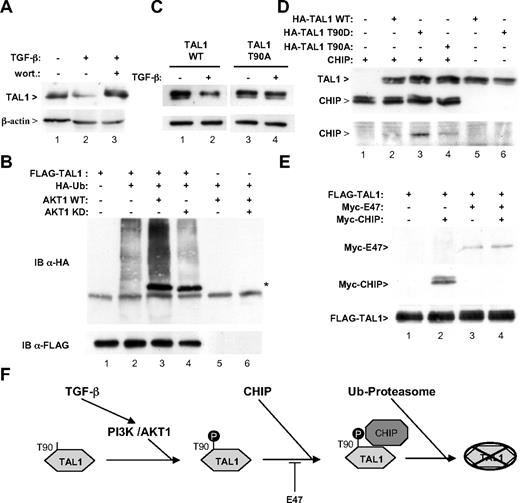
Polyubiquitylation of proteins is often induced by their phosphorylation. Because AKT1 phosphorylates TAL1 at threonine 90,13 and because TGF-β treatment is known to activate this kinase,14-17 we tested whether it might be involved in TAL1 degradation. We observed that wortmannin completely blocks the effect of TGF-β treatment on TAL1 (Figure 2A). Correspondingly, overexpression of AKT1 stimulated TAL1 polyubiquination, whereas expression of a kinase-dead mutant did not activate, and even reduced, this process (Figure 2B lanes 3 and 4) although it interacted with TAL1 as wild-type AKT1. A mutation of the AKT1 phosphorylation site of TAL1 also rendered this protein resistant to TGF-β–induced degradation (Figure 2C). These data strongly supported the notion that TAL1 phosphorylation by AKT1 triggers its polyubiquitylation and degradation. As Nie et al have recently established a role of the E3 ubiquitin ligase CHIP in the NOTCH-induced degradation of TAL1,21 we tested whether it might also be involved in the effect of TGF-β. We thus examined whether TAL1 phosphorylation could influence its interaction with CHIP. Interestingly, substitution of threonine 90 in aspartic acid (which mimics its phosphorylation13 ) was able to markedly stimulate association of TAL1 with CHIP (Figure 2D bottom panel, compare lanes 2 and 3). In contrast, substitution of threonine 90 in alanine resulted in poor binding of CHIP to TAL1 (Figure 2D bottom panel lane 4). These observations support the notion that TAL1 phosphorylation by AKT1 triggers its association with the E3 ubiquitin ligase CHIP. In agreement with this observation, both TAL1 and CHIP were detected within the nucleus of Jurkat cells (Figure S2). As Palamarchuk et al have reported that TAL1 phosphorylation by AKT1 causes its redistribution within the nucleus, this suggests that TAL1 phosphorylation and ubiquitylation takes place in the nucleus.13 As TAL1 is known to heterodimerize with the bHLH protein E47, we further tested the effect of this association on TAL1 degradation. Expression of E47 was found to strongly inhibit TAL1 polyubiquitylation (Figure S3). Likewise, coexpression of E47 abrogates the association between TAL1 and CHIP, suggesting a protective role of E47 with respect to TGF-β–induced TAL1 degradation (Figure 2E, compare lanes 2 and 4).
The PI3K/AKT1 signaling pathway is implicated in TGF-β–induced degradation of TAL1. (A) Wortmannin reverses TGF-β-induced degradation of TAL1. HeLa cells were transfected with 1 μg of pSGF-TAL1 and 10 μM wortmannin was added (lane 3) or not (lanes 1 and 2) to the culture medium 30 minutes before treatment with 10 ng/mL TGF-β1 for 9 hours (lanes 2 and 3). Cellular extracts were analyzed by immunoblot using antibodies to TAL1 (top panel) or to β-actin (bottom panel). (B) AKT1 increases TAL1 polyubiquitylation. HeLa cells were transfected with 1 μg of pSGF-TAL1 and 0.5 μg of pSG-HA-Ub, as well as 1 μg of vectors expressing HA-tagged AKT1, either wild-type (WT) or mutated in the kinase site,18 as indicated. Cell lysates were immunoprecipitated with an antibody to FLAG and immunoblot analysis was done using antibodies to HA (top panel) and to FLAG (bottom panel). (C) TAL1 T90A mutant is resistant to TGF-β–induced degradation. HeLa cells were transfected with 1 μg of pSGF-TAL1 either wild type (lanes 1 and 2) or including a change of threonine 90 to alanine (lanes 3 and 4). Cells were treated or not by TGF-β and analyzed as described for panel A. (D) TAL1 phosphorylation favors interaction with CHIP. HeLa cells were transfected with 1 μg of pRK1M/Myc-CHIP19 and HA-TAL1, either WT or including mutations T90A and T90D13 as indicated. Cell lysates were immunoprecipitated with an antibody to HA. Immunoblot analysis was done using antibodies to HA (top panel) and to Myc (bottom panel). (E) Presence of E47 prevents TAL1 interaction with CHIP. HeLa cells were transfected with 1 μg of vectors expressing pSGF-TAL1, Myc-E47,20 and Myc-CHIP as indicated. Cell lysates were immunoprecipitated with an antibody to FLAG and immunoblot analysis was done using antibodies to Myc (top panel) and to FLAG (bottom panel). (F) Schematic representation of the effect of the TGF-β/PI3K/AKT1 signaling pathway on TAL1 degradation. According to the model, TGF-β induces a PI3K/AKT1 dependent phosporylation of TAL1, which favors its interaction with CHIP, increasing TAL1 ubiquitylation and further proteolysis by the proteasome. E47, by associating with TAL1, hinders CHIP interaction.
The PI3K/AKT1 signaling pathway is implicated in TGF-β–induced degradation of TAL1. (A) Wortmannin reverses TGF-β-induced degradation of TAL1. HeLa cells were transfected with 1 μg of pSGF-TAL1 and 10 μM wortmannin was added (lane 3) or not (lanes 1 and 2) to the culture medium 30 minutes before treatment with 10 ng/mL TGF-β1 for 9 hours (lanes 2 and 3). Cellular extracts were analyzed by immunoblot using antibodies to TAL1 (top panel) or to β-actin (bottom panel). (B) AKT1 increases TAL1 polyubiquitylation. HeLa cells were transfected with 1 μg of pSGF-TAL1 and 0.5 μg of pSG-HA-Ub, as well as 1 μg of vectors expressing HA-tagged AKT1, either wild-type (WT) or mutated in the kinase site,18 as indicated. Cell lysates were immunoprecipitated with an antibody to FLAG and immunoblot analysis was done using antibodies to HA (top panel) and to FLAG (bottom panel). (C) TAL1 T90A mutant is resistant to TGF-β–induced degradation. HeLa cells were transfected with 1 μg of pSGF-TAL1 either wild type (lanes 1 and 2) or including a change of threonine 90 to alanine (lanes 3 and 4). Cells were treated or not by TGF-β and analyzed as described for panel A. (D) TAL1 phosphorylation favors interaction with CHIP. HeLa cells were transfected with 1 μg of pRK1M/Myc-CHIP19 and HA-TAL1, either WT or including mutations T90A and T90D13 as indicated. Cell lysates were immunoprecipitated with an antibody to HA. Immunoblot analysis was done using antibodies to HA (top panel) and to Myc (bottom panel). (E) Presence of E47 prevents TAL1 interaction with CHIP. HeLa cells were transfected with 1 μg of vectors expressing pSGF-TAL1, Myc-E47,20 and Myc-CHIP as indicated. Cell lysates were immunoprecipitated with an antibody to FLAG and immunoblot analysis was done using antibodies to Myc (top panel) and to FLAG (bottom panel). (F) Schematic representation of the effect of the TGF-β/PI3K/AKT1 signaling pathway on TAL1 degradation. According to the model, TGF-β induces a PI3K/AKT1 dependent phosporylation of TAL1, which favors its interaction with CHIP, increasing TAL1 ubiquitylation and further proteolysis by the proteasome. E47, by associating with TAL1, hinders CHIP interaction.
Collectively, our results establish an unexpected effect of TGF-β on TAL1 stability through AKT1 and the E3 ubiquitin ligase CHIP (Figure 2F). Recently, Nie et al have shown that NOTCH was also able to trigger degradation of TAL1 and that the SKP2 and CHIP E3 ubiquitin ligases mediate this effect.21 These authors observed that association of TAL1 with CHIP was independent of serine 300 phosphorylation.21 Our observations show that phosphorylation of TAL1 at threonine 90 stimulates CHIP binding. As NOTCH has recently been reported to induce the PI3K/AKT pathway,22 it would be interesting to test a possible role of the kinase also in NOTCH-induced TAL1 degradation. Both NOTCH and TGF-β signals are important for the normal differentiation and proliferation of T lymphocytes. It will be important in future studies to examine the relationship between both pathways on the control of TAL1 degradation, especially to examine whether they exert a synergistic effect.
TAL1 is also known to exert important functions in endothelial cells.23,24 It will be interesting to examine whether TGF-β can also control the stability of TAL1 in these cells. Indeed, Tang et al have reported that hypoxia was able to trigger TAL1 degradation in endothelial cells.25 The effect of hypoxia has been shown to result from phosphorylation of serine 122 by mitogen-activated protein kinase.25 From this result and our observations it can be proposed that the N-terminal part of TAL1 controls its stability.
In conclusion, our results show that TGF-β regulates TAL1 stability through polyubiquitylation and proteasomal degradation. As this regulation is maintained in leukemic cells and as TAL1 is essential for their proliferation,8 manipulating stability of this protein through TGF-β or AKT might offer interesting therapeutic approaches for the treatment of acute T-cell leukemias that abnormally express TAL1.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to E. Goillot, Y. Pekarsky, Z. Chang, and E. Perdiguero for gifts of plasmids, and to D. Mathieu for providing us with a monoclonal antibody to TAL1. We also thank E. MacIntyre, J.-P. Magaud, and E. Wattel for their help with T-ALL cells, and A. Roisin for assistance with cell culture.
This work was supported by the Comité du Rhône de la Ligue Nationale Contre le Cancer (J.-M.T. fellowship) and by the Institut National du Cancer.
Authorship
Contribution: J.-M.T. and P.J. designed and performed research, analyzed data, and wrote the manuscript; and L.L. and V.A. provided T-ALL cells treated by TGF-β and critically read the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Pierre Jalinot, Laboratoire de Biologie Moléculaire de la Cellule, UMR 5239, École Normale Supérieure de Lyon, 46 Allée d'Italie, 69364 Lyon cedex 07, France; e-mail: pjalinot@ens-lyon.fr.