Abstract
The proteasome inhibitor bortezomib is a novel anticancer drug that has shown promise in the treatment of refractory multiple myeloma. However, its clinical efficacy has been hampered by the emergence of drug-resistance phenomena, the molecular basis of which remains elusive. Toward this end, we here developed high levels (45- to 129-fold) of acquired resistance to bortezomib in human myelomonocytic THP1 cells by exposure to stepwise increasing (2.5-200 nM) concentrations of bortezomib. Study of the molecular mechanism of bortezomib resistance in these cells revealed (1) an Ala49Thr mutation residing in a highly conserved bortezomib-binding pocket in the proteasome β5-subunit (PSMB5) protein, (2) a dramatic overexpression (up to 60-fold) of PSMB5 protein but not of other proteasome subunits including PSMB6, PSMB7, and PSMA7, (3) high levels of cross-resistance to β5 subunit-targeted cytotoxic peptides 4A6, MG132, MG262, and ALLN, but not to a broad spectrum of chemotherapeutic drugs, (4) no marked changes in chymotrypsin-like proteasome activity, and (5) restoration of bortezomib sensitivity in bortezomib-resistant cells by siRNA-mediated silencing of PSMB5 gene expression. Collectively, these findings establish a novel mechanism of bortezomib resistance associated with the selective overexpression of a mutant PSMB5 protein.
Introduction
The ubiquitin proteasome system (UPS) facilitates the degradation of ubiquitin-tagged intracellular proteins, many of which play a regulatory role in cell proliferation, cell survival, and signaling processes.1-3 As such, proteasome inhibitors have been recognized as a new generation of chemotherapeutic agents and anti-inflammatory drugs.4-13 The boronic dipeptide bortezomib (PS341, Velcade) is the first proteasome inhibitor that has been approved for the treatment of refractory multiple myeloma.6,14 Bortezomib is a reversible inhibitor that targets primarily the β5-subunit (PSMB5) subunit/chymotrypsin-like activity of the 26S proteasome and to a somewhat lesser extent also caspase-like activity harbored by the β1 (PSMB6) proteasome subunit. At higher concentrations, bor-tezomib inhibits trypsin-like proteolytic activity facilitated by β2 (PSMB7) proteasome subunits.15-17 Despite promising clinical activity, some patients with multiple myeloma failed to respond to bortezomib therapy.18 Moreover, the efficacy for bortezomib may differ between tumor types.6,19-21 Whether these observations are related to common mechanisms of drug resistance frequently seen for anticancer22 or anti-inflammatory drugs23 is largely unknown. However, their characterization is of key importance as it may pave the way for the overcoming of drug resistance, thereby enhancing the efficacy of this new class of proteasome-targeted drugs.
One mode of primary resistance to bortezomib is conveyed by constitutively high levels of heat shock protein 27.24 In the context of acquired resistance, studies aimed at delineating the mechanism of acquired resistance to the tripeptidyl aldehyde proteasome inhibitor ALLN (N-acetyl-leucyl-leucyl-norleucinal) revealed 2 possible molecular mechanisms: (a) enhanced cellular efflux via the multidrug resistance (MDR) transporter P-glycoprotein (Pgp; ABCC1)25 or multidrug resistance-related protein 1 (MRP1; ABCC1)26 and (b) increased detoxification via up-regulated activity of aldo-keto reductase.27 Recent studies showed that Pgp overexpression conferred low levels (3-fold) of bortezomib resistance28 as well as to the irreversible proteasome inhibitor epoxomicin, but cells with 4- to 7-fold acquired resistance to epoxomicin did not show Pgp overexpression as a molecular basis of resistance.29 Two independent reports analyzing acquired resistance to the irreversible proteasome inhibitor NLVS (NIP-Leu3-vinyl sulfone) in mouse EL4 thymoma cells showed loss of proteasome function that was compensated by overexpression of another protease activity, tripeptidylpeptidase II.30-32 However, a more detailed characterization of these NVLS-resistant EL4 cells revealed that residual proteasome activity was sufficient to sustain cell function and viability33 so that the actual modality of resistance in these cells remains to be elucidated.
Altogether, given the lack of a profound understanding of the mechanisms of acquired resistance to bortezomib in mammalian cells, we set out to characterize human monocytic/macrophage cells with high levels of acquired bortezomib resistance after in vitro selection using a protocol of stepwise increases in bortezomib concentrations. Our findings establish that bortezomib resistance is not conferred by increased expression of MDR efflux pumps. Rather, qualitative and quantitative alterations were observed at the level of the β5 proteasome subunit; these included a PSMB5 gene mutation resulting in a substitution of a crucial amino acid (Ala49Thr) in the bortezomib-binding pocket of the β5-subunit. These bortezomib-resistant cells highly overexpressed this mutant β5-subunit and displayed a marked cross-resistance to β5-targeted cytotoxic peptides but not to other classes of therapeutic drugs. Hence, these findings establish a novel modality or bortezomib resistance associated with the selective overexpression of a structurally altered β5-subunit.
Methods
Cell culture and development of BTZ-resistant cell lines
Human monocytic/macrophage THP1 cells (ATCC, Manassas, VA) were kept as a suspension culture in RPMI-1640 medium supplemented with 5% fetal calf serum, 20 mM HEPES, 2 mM glutamine, and 100 μg/mL penicillin/streptomycin at 5% CO2 and 37°C. Cell cultures were seeded at a density of 3 × 105 cells/mL and refreshed twice weekly. Bortezomib (BTZ)–resistant THP1 cell lines were obtained by stepwise increasing extracellular concentrations of bortezomib over a period of 6 months, starting at a concentration of 2.5 nM (IC50 concentration: 3.3 nM) up to a concentration of 200 nM bortezomib. During this process, cultures were isolated that were grown in the presence of 30 nM (THP/BTZ30), 50 nM (THP1/BTZ50), 100 nM (THP1/BTZ100), and 200 nM bortezomib (THP1/BTZ200). All bortezomib-resistant cell lines retained parental doubling times (28 ± 3 hours) and morphology, and also retained their typical capacity to differentiate into adherent macrophage-like cells upon exposure to phorbol myristate acetate (PMA). To investigate the stability of the resistance phenotype, an aliquot of THP1/BTZ100 cells was cultured in the absence of bortezomib for a period of up to 6 months. These cells will be further designated as THP1/BTZ(-100) cells.
Proteasome activity in cell lysates
Chymotrypsin-like, trypsin-like, and caspase-like proteolytic activities of the proteasome were measured in freshly prepared cell lysates as described previously,34,35 with some minor modifications to the protocol. In brief, a total of 5 × 106 untreated or bortezomib-exposed THP1 cells were washed 3 times with ice-cold PBS and spun down by centrifugation (5 minutes, 250g, 4°C). Cell pellets were then resuspended in an ATP-containing lysis buffer (10 mM Tris-HCl buffer (pH 7.8) containing 5 mM ATP, 0.5 mM DTT, and 5 mM MgCl2) and kept on ice for 10 minutes. For complete lysis, cells were sonicated (MSE sonicator, amplitude 7, for 3 × 5 seconds with 20-second time intervals at 4°C) followed by centrifugation (5 minutes, 16 000g, 4°C) to remove cell debris. The supernatant was collected and protein concentration was determined using the Bio-Rad protein assay (Hercules, CA). Fluorogenic peptide substrates to measure the chymotrypsin-like, trypsin-like, and caspase-like activity were Suc-Leu-Leu-Val-Tyr-amc, Ac-Arg-Leu-Arg-amc, and Z-Leu-Leu-Glu-amc, respectively, all at a 100-μM final concentration. The substrates were incubated with 20 μg total cell protein extract in the presence or absence of specific inhibitors in a total assay volume of 200 μL. Specific inhibitors for chymotrypsin-like, caspase-like, and trypsin-like activity included bortezomib (10 nM), Ac-APnLD-al (25 μM), and leupeptin (20 μM), respectively. The release of AMC was monitored online over a 2-hour time period at 37°C with 5-minute intervals. Fluorescence was measured on a Tecan Spectra Fluor apparatus (Giessen, The Netherlands) using excitation and emission wavelengths of 360 and 465 nm, respectively. Proteolytic activity was calculated from the slopes of the linear portion of the curves. All results were expressed as percentage relative to untreated THP1/WT cells (100%).
Supplemental data
Information on reagents and antibodies, growth inhibition assays, apoptosis assays, immunoblotting (Western blotting and native gels), gel filtration, mitochondrial membrane potential assay, gene arrays, immunofluorescence microscopy, reverse-transcription–polymerase chain reaction (RT-PCR)/siRNA, sequence analysis, and statistics can be found in Document S1 and Figures S1Figure S2. Micro array analysis for resistance- and proteasome related genes in bortezomib-resistant THP1 cells (PDF, 32.4 KB)Figure S3. Expression of selected resistance, apoptosis and cell cycle proteins in THP1/WT cells and Bortezomib-resistant cells (PDF, 52 KB)Figure S4. Intracellular proteasome β5 and α7 subunit distribution in THP1/WT and THP1/BTZ200 cells (PDF, 53.2 KB)–S5, available on the Blood website; see the Supplemental Materials link at the top of the online article.
Results
Establishment of cells with acquired resistance to bortezomib
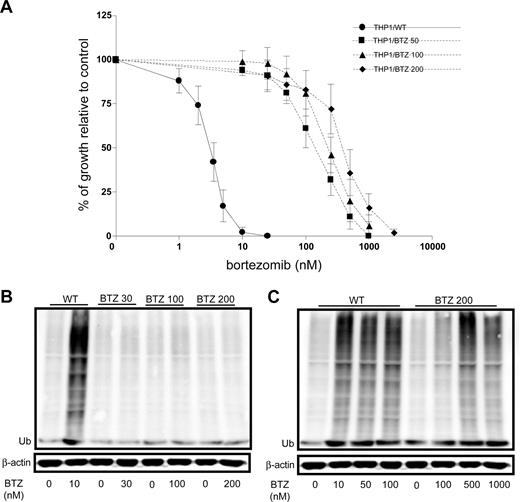
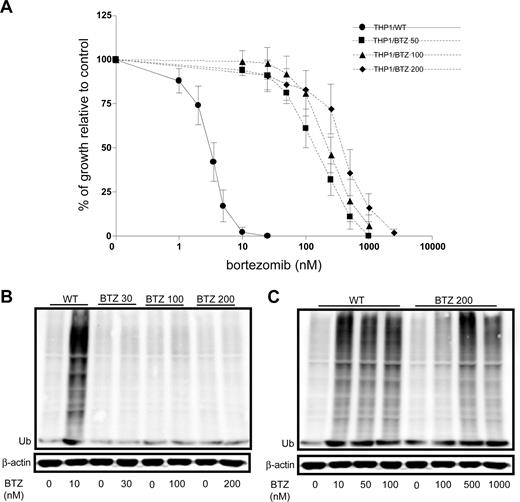
To explore the molecular basis of acquired resistance to bor-tezomib, human monocytic/macrophage THP1 cells were exposed in vitro over a period of 6 months to stepwise increasing concentrations of bortezomib representing approximately 1- to 50-fold the IC50 (2.5 nM to 200 nM). THP1 cells grown in the presence of 50 nM (THP1/BTZ50), 100 nM (THP1/BTZ100), and 200 nM (THP1/BTZ200) bortezomib were used for further characterization. Dose-response curves for bortezomib-induced cell growth inhibition revealed 45-fold (IC50: 148 ± 54 nM), 79-fold (IC50: 261 ± 71 nM), and 129-fold (IC50: 426 ± 72 nM) levels of resistance in THP/BTZ50, THP1/BTZ100, and THP1/BTZ200 cells, respectively, compared with wild-type THP/1 cells (IC50: 3.3 ± 0.6 nM; Figure 1A).
Emergence of acquired resistance to bor-tezomib and impaired bortezomib-induced accumulation of ubiquitinated proteins in bortezomib-resistant cells. (A) Dose-response curve for bortezomib-induced growth inhibition of wild-type (WT) human monocytic/macrophage THP1 cells and bortezomib (BTZ)–resistant variants THP1/BTZ50, THP1/BTZ100, and THP1/BTZ200, selected for growth in extracellular concentrations of 50 nM, 100 nM, and 200 nM bortezomib, respectively. Results depicted are the mean of 7 to 20 experiments (± SD). Drug exposure time, 72 hours. (B) Accumulation of ubiquitinated proteins in THP1/WT cells and bortezomib-resistant sublines after exposure to bor-tezomib. Bortezomib-resistant cells were allowed a 4-day drug washout period (control) after which they were exposed for 24 hours to their selective concentrations of bortezomib. THP/WT cells were exposed to 10 nM BTZ for 24 hours. (C) Accumulation of ubiquitinated proteins in THP/WT cells after 24-hour exposure to 10 to 100 nM bortezomib and for THP1/BTZ200 cells after 24-hour exposure to bortezomib concentrations beyond selective concentrations (up to 1000 nM). A representative picture of 2 separate experiments is depicted.
Emergence of acquired resistance to bor-tezomib and impaired bortezomib-induced accumulation of ubiquitinated proteins in bortezomib-resistant cells. (A) Dose-response curve for bortezomib-induced growth inhibition of wild-type (WT) human monocytic/macrophage THP1 cells and bortezomib (BTZ)–resistant variants THP1/BTZ50, THP1/BTZ100, and THP1/BTZ200, selected for growth in extracellular concentrations of 50 nM, 100 nM, and 200 nM bortezomib, respectively. Results depicted are the mean of 7 to 20 experiments (± SD). Drug exposure time, 72 hours. (B) Accumulation of ubiquitinated proteins in THP1/WT cells and bortezomib-resistant sublines after exposure to bor-tezomib. Bortezomib-resistant cells were allowed a 4-day drug washout period (control) after which they were exposed for 24 hours to their selective concentrations of bortezomib. THP/WT cells were exposed to 10 nM BTZ for 24 hours. (C) Accumulation of ubiquitinated proteins in THP/WT cells after 24-hour exposure to 10 to 100 nM bortezomib and for THP1/BTZ200 cells after 24-hour exposure to bortezomib concentrations beyond selective concentrations (up to 1000 nM). A representative picture of 2 separate experiments is depicted.
Cross-resistance profile of bortezomib-resistant cells to proteasome inhibitors and various anticancer/anti-inflammatory drugs
Bortezomib-resistant cells displayed appreciable cross-resistance to other known small (3-mer) peptide-based proteasome inhibitors (ALLN, MG132, and MG262; Table 1), although to a lower level (6- to 18-fold) than for bortezomib itself. Strikingly, very high levels of cross-resistance (up to 300-fold) were observed for the 6-mer cytotoxic peptide 4A626 that exerts a specific β5-subunit–related/chymotrypsin-like proteasome inhibitory activity (R.O., Y.G.A., R.J.S., G.J., unpublished results). Of note, no appreciable levels of cross-resistance were observed for a broad spectrum of chemotherapeutic drugs with distinct mechanisms of action, for example, methotrexate (folate antagonist), sulfasalazine (inhibitor κB kinase/NFκB inhibitor), 5-fluorouracil (fluoropyrimidine antimetabolite), chloroquine (lysosomotropic drug), bleomycin (DNA-interacting agent), gefitinib (epidermal growth factor receptor tyrosine kinase inhibitor), cisplatin (DNA interchelator), cyclosporin A (immunosuppressive drug), methylprednisolone (corticosteroid), geldanamycin (heat shock protein inhibitor), doxorubicin (DNA-interacting drug), and mitoxantrone (topoisomerase inhibitor; Table 1). The latter 2 drugs are bona fide substrates of the ATP-driven drug efflux transporters P-glycoprotein (Pgp; ABCB1), multidrug resistance-associated proteins 1-9 (MRP1-9/ABCCs), or breast cancer resistance protein (BCRP; ABCG2).22 The lack of cross-resistance to these drugs in bortezomib-resistant cells argues against a multidrug resistance (MDR) phenotype. This notion was further corroborated by the observation that parental mRNA levels of Pgp, MRP1-9, and BCRP were retained in bortezomib-resistant cells (data not shown) and Western blot experiments (Figure S1) showing no differential expression of Pgp, MRP1-6, and BCRP in bortezomib-resistant cells versus parental THP1/WT cells. Finally, bortezomib-resistant cells lacked cross-resistance to AAF-cmk, an inhibitor of the proteolytic system tripeptidylpeptidase II.33 Together, these results demonstrate that cross-resistance of bortezomib-resistant cells is restricted to (peptide) drugs that primarily target the proteasome's β5-subunit.
To further characterize the bortezomib-resistant cells, and to gain insight whether multiple mechanisms of resistance may contribute to the bortezomib-resistant phenotype, microarray ana-lysis was performed to assess differential gene expression in control THP1/WT cells versus THP1/BTZ30 cells, THP1/BTZ100 cells, and its subline THP1/BTZ(-100) grown in the absence of BTZ for 6 months. The entire data set of the test series is available at the GEO database, accession number GSE11771.36 Preliminary evaluation of expression of genes specifically involved in drug metabolism and resistance (n = 101), apoptosis (n = 59), and cell cycling (n = 76) showed no marked and/or robust alterations (< 2- or > 2-fold) that could be associated with resistance (Figure S2). Consistently, Western blot analysis for specific proteins in these gene categories (eg, Hsp27, Hsp90, XIAP, and p21) showed no marked changes in bortezomib-resistant cells compared with bortezomib-sensitive THP1/WT cells (Figure S3). Strikingly, however, approximately 40% of genes representing the various proteasome subunits, in particular β-subunits, showed a markedly (≥ 2-fold) increased expression level (Figure S2). Only 2 proteasome subunit-related genes were down-regulated in expression (≤ 2-fold): the immunoproteasome subunits β5i and β1i. Of additional interest, no alterations were observed in expression levels of genes encoding ubiquitin-conjugating enzymes, ubiquitin-specific proteases, or ubiquitin C-terminal hydrolases. Finally, up-regulated expression of proteasome subunit genes in bortezomib-resistant cells appeared to be transient as gene expression normalized when THP1/BTZ100 cells were grown in the absence of bortezomib for 6 months (Figure S2). Together, gene array analyses further support the notion that alterations in proteasome subunit composition or function may be dominantly involved in the bortezomib-resistant phenotype.
Diminished accumulation of ubiquitinated proteins in bortezomib-resistant cells
A characteristic feature of proteasome inhibition is the accumulation of ubiquitinated proteins that provokes loss of mitochondrial membrane potential and apoptosis.33,37 Consistently, marked accumulation of ubiquitinated proteins was observed in wild-type cells exposed to bortezomib but not in bortezomib-resistant cells at their selective concentration of 30 to 200 nM (Figure 1B). However, the latter cells showed clear accumulation of ubiquitinated proteins if exposed to concentrations of bortezomib above the selective concentrations (500-1000 nM; Figure 1C). Thus, bortezomib-resistant cells have retained their capacity to accumulate ubiquitinated proteins, except that this process is initiated at markedly higher concentrations of bortezomib consistent with the resistance factor for bortezomib.
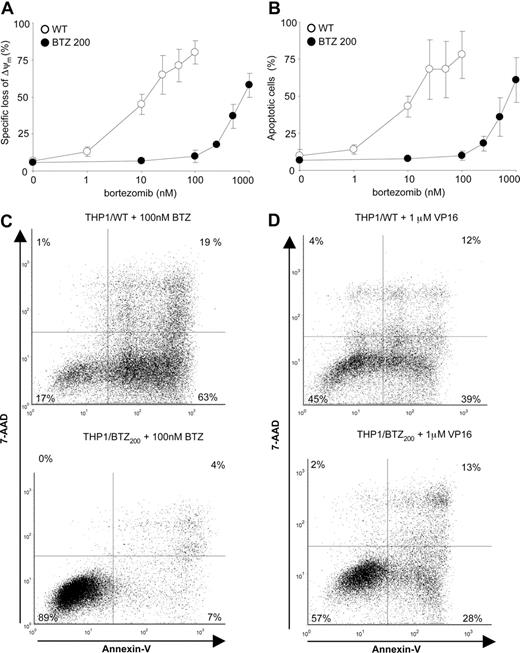
Impaired bortezomib-induced loss of mitochondrial membrane potential and induction of apoptosis in bortezomib-resistant cells
The shift in bortezomib-induced accumulation of ubiquitinated proteins in bortezomib-resistant cells paralleled the shift in bortezomib-induced loss of mitochondrial membrane potential (Figure 2A) and induction of apoptosis (Figure 2B,C). For comparison, the anticancer drug and topoisomerase II inhibitor etoposide (VP16) was equally effective in inducing apoptosis in wild-type as well as bortezomib-resistant cells (Figure 2D). These results indicate that bortezomib-resistant cells have retained their capacity to undergo apoptosis, consistent with the lack of alterations in gene expression of apoptosis-related genes (Figure S2), but the ability to undergo apoptosis is specifically impaired for proteasome inhibitor type of drugs.
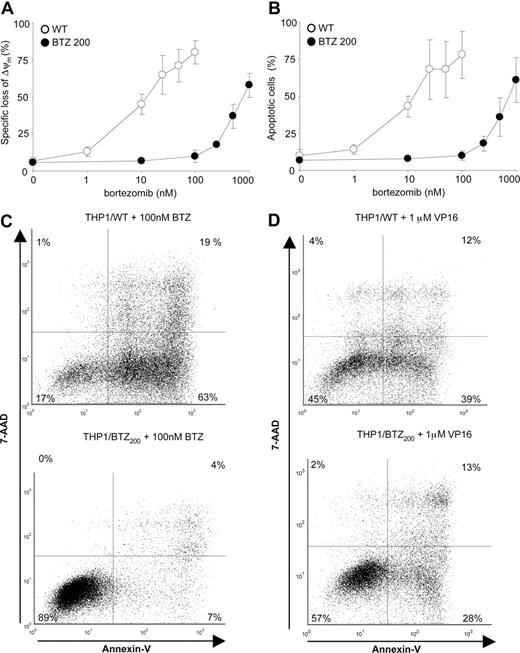
Differential induction of apoptosis by bor-tezomib and VP16 in bortezomib-resistant cells. (A) Loss of mitochondrial membrane potential (Δψm) after 24-hour exposure of THP1/WT cells and THP1/BTZ200 cells to a concentration range of bortezomib. (B) Induction of apoptosis (annexin V–positive cells) in THP1/WT cells and THP1/BTZ200 cells after 24-hour exposure to a concentration range of bortezomib. (C) A representative flow cytometric picture of apoptosis induction (annexinV/7-AAD staining) after 24-hour incubation of THP1/WT cells and THP1/BTZ200 cells with 100 nM BTZ. (D) A representative flow cytometric picture of apoptosis induction (annexin-V/7-AAD staining) after 48-hour incubation of THP1/WT cells and THP1/BTZ200 cells with 1 μM VP-16/etoposide. All results present the means (± SD) for 3 independent experiments.
Differential induction of apoptosis by bor-tezomib and VP16 in bortezomib-resistant cells. (A) Loss of mitochondrial membrane potential (Δψm) after 24-hour exposure of THP1/WT cells and THP1/BTZ200 cells to a concentration range of bortezomib. (B) Induction of apoptosis (annexin V–positive cells) in THP1/WT cells and THP1/BTZ200 cells after 24-hour exposure to a concentration range of bortezomib. (C) A representative flow cytometric picture of apoptosis induction (annexinV/7-AAD staining) after 24-hour incubation of THP1/WT cells and THP1/BTZ200 cells with 100 nM BTZ. (D) A representative flow cytometric picture of apoptosis induction (annexin-V/7-AAD staining) after 48-hour incubation of THP1/WT cells and THP1/BTZ200 cells with 1 μM VP-16/etoposide. All results present the means (± SD) for 3 independent experiments.
Marked overexpression of proteasome subunit β5 but not chymotrypsin-like proteasomal activity in bortezomib-resistant cells
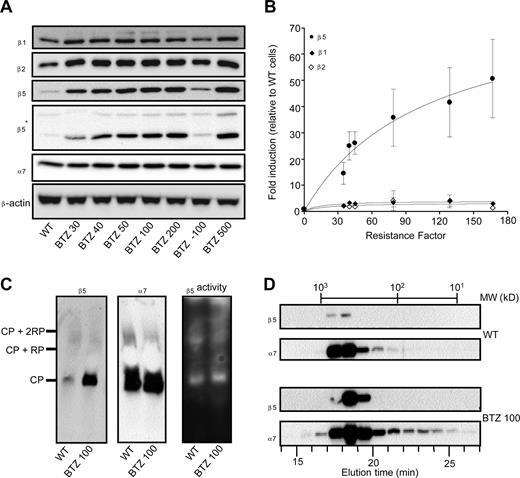
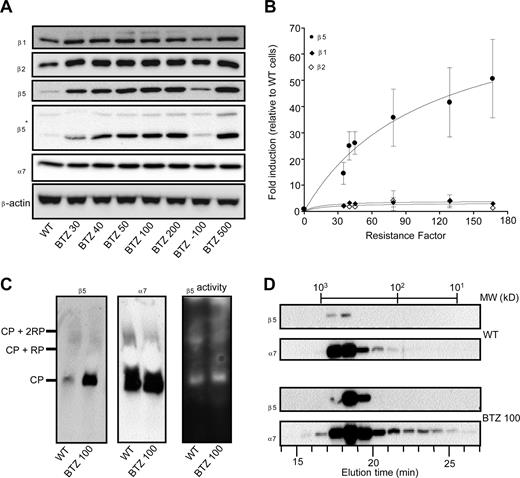
Because the cross-resistance data (Table 1) were consistent with specific alterations at the level of proteasome subunit expression and/or catalytic activity, the expression pattern of individual β1-, β2-, and β5-subunits was determined in wild-type and drug-resistant cells grown at their selective bortezomib concentrations. Furthermore, to explore the stability of the bortezomib-resistance phenotype, we also examined a subline of THP1/BTZ100 that was grown in the absence of bortezomib for 6 months (termed THP1/BTZ(-100)). Proteasome β1 and β2-subunit expression was only slightly increased (< 2-fold), whereas proteasome α7-subunit expression was not significantly altered in the bortezomib-resistant cells (Figure 3A). In contrast, β5 protein levels, relatively barely detectable in parental cells, were dramatically increased (up to 60-fold) in bortezomib-resistant cells (Figure 3A). This observation was confirmed using 2 different antibodies recognizing different epitopes within the β5 protein. This dramatic increase in β5 expression was proportional to the gradually increasing concentrations of bortezomib during the stepwise selection. Notably, over a concentration range (up to 50 nM bortezomib) where bortezomib primarily inhibits chymotrypsin-like proteasome activity, β5 overexpression parallels the extent of bortezomib resistance (Figure 3B). Apart from Western blot analysis, we also explored whether β5 overexpression was associated with the 20S core particle of the proteasome by performing native gel electrophoresis analysis. Results depicted in Figure 3C further demonstrate an increased expression of β5 in resistant cells compared with parental cells; but all of this increased expression was confined to the 20S proteasome complex rather than being present as unassembled “free” subunits. As a control, expression of subunit α7 remained unchanged, as did the overall proteolytic activity in both parental and resistant cells using suc-LLVY-amc as a substrate (Figure 3C). This finding was further corroborated by gel filtration experiments (Figure 3D) that demonstrated that elevated expression of β5 was observed in bortezomib-resistant cells that eluted from the column solely as a high-molecular-weight fraction of approximately 600 to 700 kDa protein, consistent with the 20S core particle's molecular weight. In accord with data from Figure 3A,C, α7-subunit expression was largely unaltered between parental and bortezomib-resistant cells, although its presence was not exclusively restricted to high-molecular-weight fractions, but also showed up in lower molecular weight fractions, possibly even as “free” subunits. Consistent with these experiments, immunohistochemical analysis (Figure S4) also confirmed the colocalization of β5 and α7 in THP1/BTZ cells.
Selective induction of proteasome β5-subunits in bortezomib-resistant cells. (A) Protein expression of proteasome β5-, β2-, and β1-subunits and α7-subunits in wild-type and bortezomib-resistant THP1 sublines. THP1/BTZ(-100) represents a subline of THP1/BTZ100 that was grown in the absence of bor-tezomib for 6 months. Note: 2 different sources of β5 antibodies were used: Biomol (PW8895; Plymouth Meeting, PA) and 20S X from Novus Biologicals (Littleton, CO), the latter indicated by an asterisk (*). (B) Induction of proteasome subunits β5, β2, and β1 in relation to resistance factors to bortezomib for the selected panel of bortezomib-resistant THP1 cells. Densitometry results are presented as the mean (± SD) of 4 separate experiments. (C) Native gel electrophoresis of crude cell extracts of THP1/WT cells and THP1/BTZ100 cells subsequently analyzed for β5- and α7-subunit expression (left panels) and catalytic activity for the substrate Suc-LLVY-AMC (right panel). (D) Gel filtration of crude extracts of THP1/WT cells and THP1/BTZ100 cells via a high-performance liquid chromatography (HPLC)–linked Superdex 200 HR 10/30 column (Supelco, Bellefonte, CA). Proteins were eluted by washing the column with 20 mM Tris-HCl, pH 7.5, 5 mM ATP, and 120 mM NaCl at a flow rate of 0.4 mL/min. Fractions were collected every minute and subject to Western blot analysis for β5 and α7. The column was calibrated with a mixture of purified proteins in the MW range of 16.6 kDa to 669 kDa.
Selective induction of proteasome β5-subunits in bortezomib-resistant cells. (A) Protein expression of proteasome β5-, β2-, and β1-subunits and α7-subunits in wild-type and bortezomib-resistant THP1 sublines. THP1/BTZ(-100) represents a subline of THP1/BTZ100 that was grown in the absence of bor-tezomib for 6 months. Note: 2 different sources of β5 antibodies were used: Biomol (PW8895; Plymouth Meeting, PA) and 20S X from Novus Biologicals (Littleton, CO), the latter indicated by an asterisk (*). (B) Induction of proteasome subunits β5, β2, and β1 in relation to resistance factors to bortezomib for the selected panel of bortezomib-resistant THP1 cells. Densitometry results are presented as the mean (± SD) of 4 separate experiments. (C) Native gel electrophoresis of crude cell extracts of THP1/WT cells and THP1/BTZ100 cells subsequently analyzed for β5- and α7-subunit expression (left panels) and catalytic activity for the substrate Suc-LLVY-AMC (right panel). (D) Gel filtration of crude extracts of THP1/WT cells and THP1/BTZ100 cells via a high-performance liquid chromatography (HPLC)–linked Superdex 200 HR 10/30 column (Supelco, Bellefonte, CA). Proteins were eluted by washing the column with 20 mM Tris-HCl, pH 7.5, 5 mM ATP, and 120 mM NaCl at a flow rate of 0.4 mL/min. Fractions were collected every minute and subject to Western blot analysis for β5 and α7. The column was calibrated with a mixture of purified proteins in the MW range of 16.6 kDa to 669 kDa.
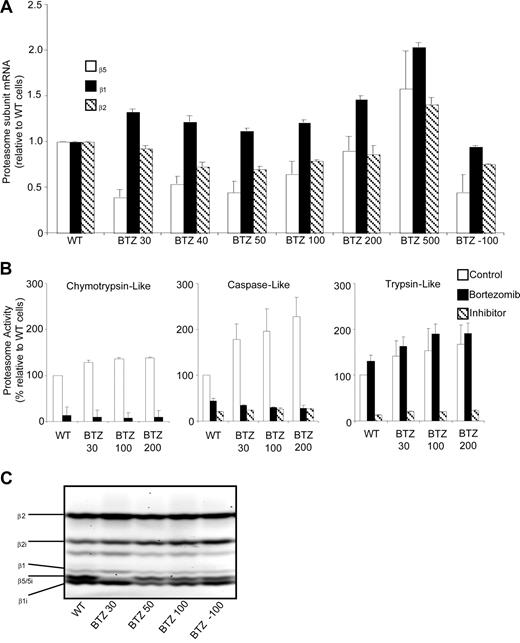
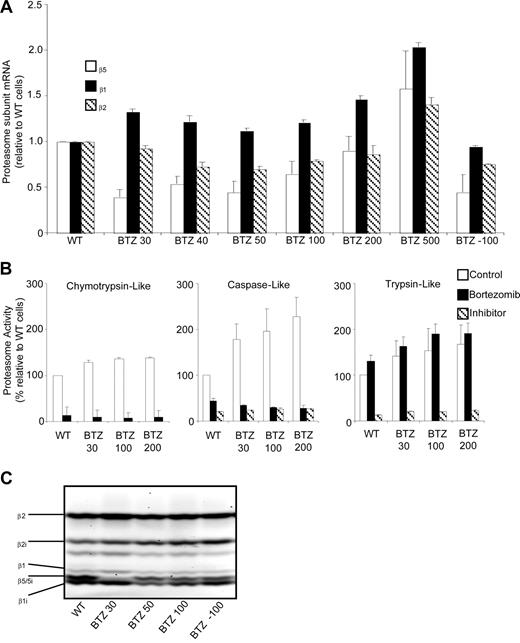
In contrast to β5 protein levels, β5 mRNA levels in bortezomib-resistant cells were only marginally increased at the highest selection dose (Figure 4A), suggesting that the induction of β5 protein most likely is effectuated at the posttranscriptional level. As for the β5 transcript, β2 and β1 mRNA levels were only modestly elevated in cells with the highest bortezomib-resistance level (Figure 4A).
Proteasome β5-, β2-, and β1-subunit mRNA levels and proteasome subunit–related catalytic activity in wild-type and bortezomib-resistant cells. (A) mRNA levels for proteasome β5-, β2-, and β1-subunits in selected variants of bortezomib-resistant THP1 cells relative to THP1/WT cells. mRNA levels were quantified using β-glucuronidase (GUS) as reference gene and depicted relative to THP1/WT cells. (B) Chymotrypsin-like, caspase-like, and trypsin-like proteasomal activities assayed with specific fluorogenic peptide substrates in cell extracts of THP1/WT, THP1/BTZ30, THP1/BTZ100, and THP1/BTZ200 cells after a 4-day drug washout period (control) and after 24-hour incubation with 10 nM bor-tezomib (for THP/WT) and selective concentrations of 30 nM, 100 nM, and 200 nM bortezomib for the indicated bortezomib-resistant THP1 sublines. Controls for selective inhibition of caspase-like activity and trypsin-like activity included Ac-APnLD (25 μM) and leupeptin (20 μM), respectively. All results represent the mean (± SD) of 3 independent experiments. (C) Activity labeling of constitutive and immunoproteasome β-subunits in intact THP1/WT and selected bortezomib-resistant THP/BTZ cells using bodipyFL-Ahx3L3VS affinity probe.
Proteasome β5-, β2-, and β1-subunit mRNA levels and proteasome subunit–related catalytic activity in wild-type and bortezomib-resistant cells. (A) mRNA levels for proteasome β5-, β2-, and β1-subunits in selected variants of bortezomib-resistant THP1 cells relative to THP1/WT cells. mRNA levels were quantified using β-glucuronidase (GUS) as reference gene and depicted relative to THP1/WT cells. (B) Chymotrypsin-like, caspase-like, and trypsin-like proteasomal activities assayed with specific fluorogenic peptide substrates in cell extracts of THP1/WT, THP1/BTZ30, THP1/BTZ100, and THP1/BTZ200 cells after a 4-day drug washout period (control) and after 24-hour incubation with 10 nM bor-tezomib (for THP/WT) and selective concentrations of 30 nM, 100 nM, and 200 nM bortezomib for the indicated bortezomib-resistant THP1 sublines. Controls for selective inhibition of caspase-like activity and trypsin-like activity included Ac-APnLD (25 μM) and leupeptin (20 μM), respectively. All results represent the mean (± SD) of 3 independent experiments. (C) Activity labeling of constitutive and immunoproteasome β-subunits in intact THP1/WT and selected bortezomib-resistant THP/BTZ cells using bodipyFL-Ahx3L3VS affinity probe.
We next examined the possible role that these quantitative alterations in β5-subunit composition may bear on the various proteolytic activities of the proteasome. Proteasomal proteolytic activities represented by chymotrypsin-like activity, caspase-like activity, and trypsin-like activity were measured using fluorogenic peptide substrates in cell extracts of parental cells incubated for 24 hours in the presence or absence of 10 nM bortezomib. Bortezomib-resistant cells were tested after a 4-day drug-free period (control), and after 24-hour incubation at their selective bortezomib concentration (Figure 4B). After 4 days of incubation in drug-free medium, bortezomib-resistant cells had a small (1.3- to 1.4-fold) increase in chymotrypsin-like activity compared with wild-type cells. Under bortezomib-selective conditions (30, 100, or 200 nM), residual chymotrypsin-like activity was reduced to 8% to 10% of drug-free controls, as observed for parental cells exposed to 10 nM bortezomib. After drug washout of bortezomib-resistant cells, caspase-like proteasome activity was significantly increased (1.8- to 2.3-fold; P < .001) over parental cells. In the presence of bortezomib, caspase-like activity was reduced to 26% to 35% of wild-type levels. Finally, basal trypsin-like activity was 1.4- to 1.7-fold (P < .01) increased in bortezomib-resistant cells compared with wild-type cells. In the presence of bortezomib, trypsin-like activity was stimulated by another 20% to 40% both in bortezomib-resistant cell lines and wild-type cells (Figure 4B). These results indicate that the qualitative characteristics of proteasomal inhibition by bortezomib were retained in bortezomib-resistant cells, whereas quantitatively, a marked up-regulation of β5-subunit protein did not lead to a β5-subunit–related increase in chymotrypsin-like proteasome activity.
Finally, probing of active proteasome subunits was performed using the bodipyFL-Ahx3L3VS affinity labeling reagent.38 Labeling of constitutive β5 and its immunoproteasome counterpart β5i was readily observed in parental THP1 cells (Figure 4C), but markedly inhibited at relatively low levels of bortezomib resistance in THP1/BTZ30 cells. Interestingly, probing of active β5 and β5i was partly restored in THP1 cells with higher levels of bortezomib resistance including THP1/BTZ50 and THP1/BTZ100 cells, thus underscoring the fact that proteasome activity is retained.
Stability of the bortezomib-resistance phenotype
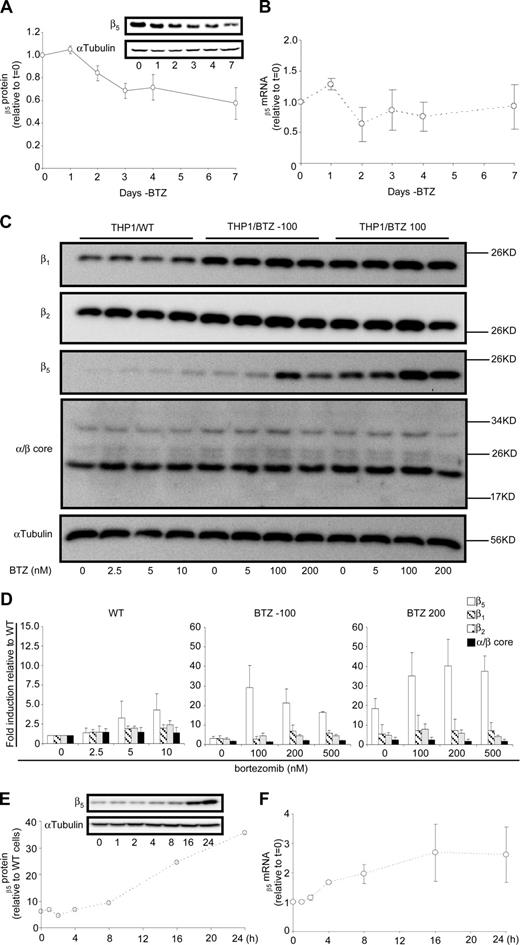
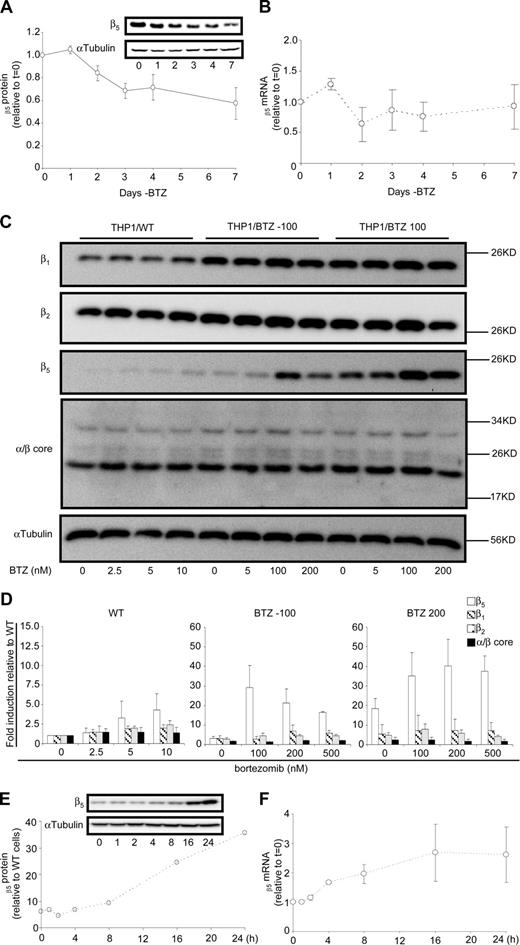
To investigate the stability of the resistance phenotype and expression of β5-subunit in the absence of bortezomib, resistant cells were transferred to bortezomib-free medium for a period of 7 days and showed a gradual decline of β5 expression to 40% of the original levels (Figure 5A); whereas, no substantial alterations were observed in β5-subunit mRNA levels (Figure 5B). Interestingly, however, Figure 3A demonstrated that β5-subunit expression was further reduced in THP1/BTZ(-100) cells that were cultured in bortezomib-free medium for 6 months, although they had retained a more than 35-fold resistance level for bortezomib (not shown). This prompted us to investigate whether β5-subunit expression could be rapidly reinduced upon bortezomib reexposure. Indeed, when THP1/WT, THP1/BTZ(-100), and THP1/BTZ100 cells were exposed to a range of bortezomib concentrations (1-3 × IC50) for 24 hours, a marked and selective induction of β5-subunit expression was noted for THP1/BTZ(-100) cells and to a lesser extent for THP1/BTZ100 cells (Figure 5C,D). Fine-tuning the 0- to 24-hour time course of β5 induction revealed that within 8 to 16 hours after bortezomib exposure, β5-subunit expression was markedly induced in THP1/BTZ(-100) cells (Figure 5E) and was accompanied by a 2- to 3-fold induction of β5 mRNA levels (Figure 5F). Collectively, these results suggest that once bortezomib resistance has been provoked after chronic exposure, the drug resistance phenotype not only can remain dormant for long-term periods when cells are not exposed to bortezomib, but it would also be rapidly revived upon reexposure to bortezomib.
Dynamics of proteasome subunit expression in bortezomib-resistant cells after exposure and removal of bortezomib. (A) Effect of 1- to 7-day removal of bortezomib-selective pressure from THP1/BTZ100 cells on proteasome subunit β5 protein expression and (B) β5 proteasome subunit mRNA expression. Results are the means (± SD) of 3 separate experiments. (C) Protein expression of β1-, β2-, β5- and α/β-core proteasome subunits in THP1/WT cells, THP1/BTZ(-100) cells, and THP1/BTZ200 cells after 4-day bortezomib washout (control/0) and 24-hour exposure to the indicated concentrations of bortezomib. (D) Protein expression of β1, β2, β5- and α/β-core proteasome subunits for THP1/WT cells, THP1/BTZ(-100) cells, and THP1/BTZ200 cells after a 4-day bortezomib washout period (control) and after 24-hour incubation with the indicated concentrations of bor-tezomib. Results of scanning of protein band intensities are presented as the mean ± SD of 3 separate experiments. (E) Effect of short-term (0-24 hours) exposure of THP1/BTZ(-100) cells to 100 nM bortezomib on proteasome subunit β5 protein expression and (F) β5 proteasome subunit mRNA expression. Results are the means of duplicate experiments (E) and 3 separate experiments (± SD; F).
Dynamics of proteasome subunit expression in bortezomib-resistant cells after exposure and removal of bortezomib. (A) Effect of 1- to 7-day removal of bortezomib-selective pressure from THP1/BTZ100 cells on proteasome subunit β5 protein expression and (B) β5 proteasome subunit mRNA expression. Results are the means (± SD) of 3 separate experiments. (C) Protein expression of β1-, β2-, β5- and α/β-core proteasome subunits in THP1/WT cells, THP1/BTZ(-100) cells, and THP1/BTZ200 cells after 4-day bortezomib washout (control/0) and 24-hour exposure to the indicated concentrations of bortezomib. (D) Protein expression of β1, β2, β5- and α/β-core proteasome subunits for THP1/WT cells, THP1/BTZ(-100) cells, and THP1/BTZ200 cells after a 4-day bortezomib washout period (control) and after 24-hour incubation with the indicated concentrations of bor-tezomib. Results of scanning of protein band intensities are presented as the mean ± SD of 3 separate experiments. (E) Effect of short-term (0-24 hours) exposure of THP1/BTZ(-100) cells to 100 nM bortezomib on proteasome subunit β5 protein expression and (F) β5 proteasome subunit mRNA expression. Results are the means of duplicate experiments (E) and 3 separate experiments (± SD; F).
Identification of a mutation in the PSMB5 gene in THP1/BTZ cells
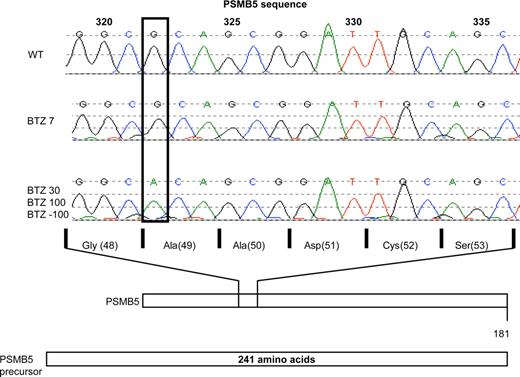
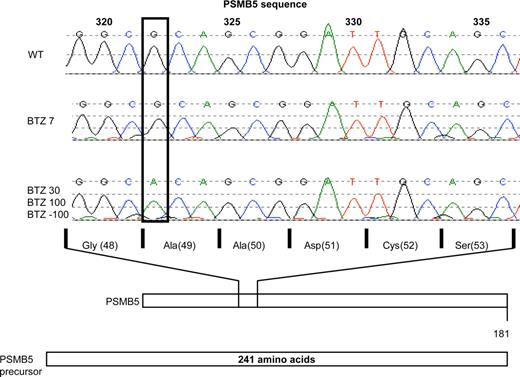
Given the stable and long-term retention of the bortezomib-resistance phenotype, along with the dominant involvement of the β5-subunit, we explored whether a genetic alteration in the PSMB5 gene could have contributed to the onset of acquired resistance to bortezomib. To this end, we sequenced part of exon 2 of the PSMB5 gene that encodes for the highly conserved binding pocket region for proteasome inhibitors within the β5 protein. In THP1 cells, selected for a very low level of acquired resistance to bortezomib, that is, 7 nM (THP1/BTZ7), no genetic alterations could be observed (Figure 6). However, in THP1 cells displaying higher levels of bortezomib resistance including THP1/BTZ30 and THP1/BTZ100 cells, a single G to A nucleotide shift was identified at position 322 of the PSMB5 gene. In the mature and functional β5-subunit protein, this mutation introduces an Ala to Thr substitution at amino acid 49 (Figure 6). We further established that the Ala49Thr mutation was retained in THP1/BTZ(-100) cells (Figure 6). Based on the fact that Ala49 resides in the highly conserved substrate/inhibitor binding domain of the β5-subunit, this homozygous mutation is likely to contribute to the bortezomib-resistance phenotype.
Analysis of PSMB5 gene mutations in bor-tezomib-resistant THP1 cells. Sequencing of PSMB5 gene exon 2 in THP1/WT cells and various BTZ-resistant sublines: THP1/BTZ7, THP1/BTZ30, THP1/BTZ100, and THP1/BTZ(-100). Depicted is the single nucleotide shift (G→A) at nucleotide position 322 in THP1/BTZ30, THP1/BTZ100, and THP1/BTZ(-100) along with the corresponding change in a single amino acid substitution (Ala49Thr) within the mature PSMB5/β5 protein.
Analysis of PSMB5 gene mutations in bor-tezomib-resistant THP1 cells. Sequencing of PSMB5 gene exon 2 in THP1/WT cells and various BTZ-resistant sublines: THP1/BTZ7, THP1/BTZ30, THP1/BTZ100, and THP1/BTZ(-100). Depicted is the single nucleotide shift (G→A) at nucleotide position 322 in THP1/BTZ30, THP1/BTZ100, and THP1/BTZ(-100) along with the corresponding change in a single amino acid substitution (Ala49Thr) within the mature PSMB5/β5 protein.
SiRNA-dependent silencing of β5 induction in bortezomib-resistant cells reverses bortezomib sensitivity and induces apoptosis
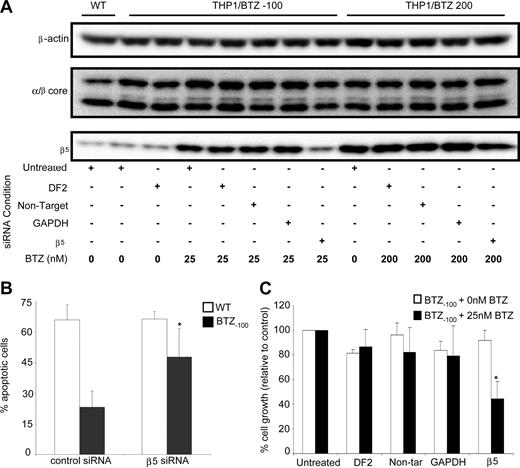
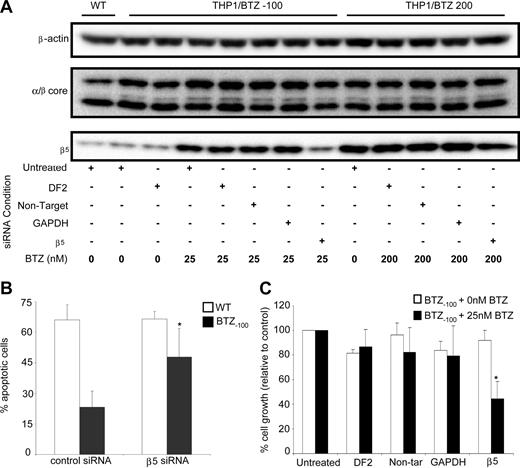
Finally, based on data in Figure 5, we determined whether prevention of β5-subunit induction would lead to restoration of bortezomib sensitivity. To this end, the consequences of siRNA-based silencing of β5 expression were explored in THP1/WT, THP1/BTZ(-100), and THP1/BTZ200 cells. In THP1/BTZ(-100) cells, the β5-siRNA–dependent prevention of β5-subunit up-regulation after bortezomib exposure (Figure 7A) was accompanied by both a significantly higher fraction of apoptotic cells (Figure 7B) as well as increased bortezomib sensitivity (Figure 7C). For THP1/BTZ200 cells with constitutively high levels of β5 expression (Figure 3A), β5 siRNA–dependent silencing also repressed β5 expression (Figure 7A), but not to a level that compromised bortezomib resistance (results not shown). Collectively, these results further underscore the involvement of proteasome β5-subunit overexpression in conferring bortezomib resistance and provide a targeted strategy to reverse bortezomib resistance.
SiRNA-mediated silencing of proteasome β5-subunit induction restores bortezomib sensitivity and induces apoptosis in bortezomib-resistant cells. (A) β5-siRNA–induced silencing of β5-protein expression in THP1/BTZ(-100) and THP1/BTZ200 cells after 24-hour preincubation with transfection medium (DF2), control siRNA constructs (nontarget/GAPDH), and a β5-specific siRNA construct (transduction efficiency: > 85% based on siGlo+ cells by flow cytometry). After transfection, cells were incubated (24-48 hours) with the indicated concentrations of bortezomib. Protein expressions of α/β-core and β-actin are shown as controls. A representative picture of 3 separate experiments is depicted. (B) Bortezomib-induced apoptosis in THP1/WT, THP1/BTZ(-100), and THP1/BTZ200 cells after siRNA-induced prevention of induction of β5-subunit expression as described in panel A. Bor-tezomib exposure time: 24 hours. Bortezomib concentrations used: 25 nM for THP1/WT and THP1/BTZ(-100) cells. Means of 4 separate experiments (± SD) are shown. *P < .01. (C) Bortezomib-induced cell growth inhibition of THP1/BTZ(-100) cells after prevention of induction of β5-subunit expression by siRNA silencing as described in panel A and 24-hour exposure to 25 nM bortezomib. Means of 3 separate experiments (± SD) are shown. *P < .01.
SiRNA-mediated silencing of proteasome β5-subunit induction restores bortezomib sensitivity and induces apoptosis in bortezomib-resistant cells. (A) β5-siRNA–induced silencing of β5-protein expression in THP1/BTZ(-100) and THP1/BTZ200 cells after 24-hour preincubation with transfection medium (DF2), control siRNA constructs (nontarget/GAPDH), and a β5-specific siRNA construct (transduction efficiency: > 85% based on siGlo+ cells by flow cytometry). After transfection, cells were incubated (24-48 hours) with the indicated concentrations of bortezomib. Protein expressions of α/β-core and β-actin are shown as controls. A representative picture of 3 separate experiments is depicted. (B) Bortezomib-induced apoptosis in THP1/WT, THP1/BTZ(-100), and THP1/BTZ200 cells after siRNA-induced prevention of induction of β5-subunit expression as described in panel A. Bor-tezomib exposure time: 24 hours. Bortezomib concentrations used: 25 nM for THP1/WT and THP1/BTZ(-100) cells. Means of 4 separate experiments (± SD) are shown. *P < .01. (C) Bortezomib-induced cell growth inhibition of THP1/BTZ(-100) cells after prevention of induction of β5-subunit expression by siRNA silencing as described in panel A and 24-hour exposure to 25 nM bortezomib. Means of 3 separate experiments (± SD) are shown. *P < .01.
Discussion
This study provides several lines of evidence for the proteasome involvement, that of the β5-subunit in particular, in the acquisition of bortezomib resistance in THP1 cells: (1) a mutation in the PSMB5 gene involving an Ala49Thr substitution in the highly conserved substrate/inhibitor binding domain of the β5-subunit, (2) selective overexpression of the mutant β5-subunit protein paralleled bortezomib-resistance levels in THP1/BTZ cells, (3) rapid and marked induction of β5 in THP1/BTZ(-100) cells after exposure to bortezomib, (4) siRNA-guided silencing of β5-subunit gene expression restored bortezomib sensitivity and induced apoptosis, and (5) bortezomib-resistant cells displayed marked cross-resistance to other peptide-based proteasome inhibitors, in particular those that specifically target the β5-subunit.
Crystallography data from yeast and mammalian proteasomes39,40 indicated that Ala49 is implicated in the efficient binding of bortezomib in the substrate/inhibitor binding pocket of the β5-subunit. Moreover, Ala49 is a highly conserved residue among many prokaryotic and mammalian species. As such, substitution of Ala49 for Thr containing a neutral polar side chain is likely to have consequences for efficiency of (reversible) binding of bortezomib and other peptide-based proteasome inhibitors. Prototypically this may be exemplified by the 6-mer peptide 4A6 that selectively targets β5 (but not subunits β1 and β2) for which up to more than 250-fold level of cross-resistance was observed in the bortezomib-resistant cells. The finding that cross-resistance levels for ALLN, MG132, and MG262 were lower than the resistance factors for bortezomib may be consistent with the fact that these proteasome inhibitors have additional targets apart from β5, β2, and β1 proteasome subunits, for example, lysosomal proteases, which may not be affected in bortezomib-resistant cells.9,15,20,38 The potential impact of Ala49 mutations in conferring bortezomib resistance may be further supported by recent preliminary observations by Lu et al41 showing the same PSMB5 mutation in bortezomib-resistant human lymphoblastic Jurkat T cells.
In keeping with the notion of a prominent role of the β5 proteasome subunit in conferring high levels of resistance to bortezomib, other types of molecular mechanisms previously reported to influence bortezomib activity did not seem to apply for bortezomib-resistant THP1 cells. No evidence for extensive metabolism (oxidative deboronation) of bortezomib by cytochrome P450 reactions42 could be obtained from gene array analysis of CytP450 enzymes. A putative role for MDR proteins in bortezomib-resistant cells could be ruled out both by the lack of cross-resistance to prototypical MDR substrates (doxorubicin and mitoxantrone) and by the lack of up-regulation of MDR proteins typically involved in drug resistance. Consistently, Minderman et al28 and our laboratory (R.O., R.J.S., G.J., unpublished observations, April 2008) showed that low levels (< 3-fold) of resistance to bortezomib were detected only in mammalian cells expressing high levels of Pgp, and not by other MDR drug efflux transporters such as MRP1-6 or BCRP. In fact, none of these MDR transporters were found to be overexpressed in bortezomib-resistant THP1 cells. Furthermore, up-regulation of another proteolytic system, tripeptidylpeptidase II, to compensate for proteasome inhibition by irreversible proteasome inhibitors,30-32 was ruled out based on the lack of cross-resistance to the TPP II inhibitor, AAF-cmk.33 Finally, potential loss of bortezomib activity after short drug exposures due to constitutive or transient induction of heat shock proteins (Hsp's)24,43 was excluded in the present chronic exposure model as judged from the lack of cross-resistance to the potent Hsp inhibitor geldanamycin44 and unaltered hsp27 expression in bortezomib-resistant cells (Figure S3).
Up-regulation of target enzymes is a common mode of resistance to several types of chemotherapeutic drugs.45,46 In this context, effects on proteasomal β5-subunits as the primary target for bortezomib may not be unexpected. In fact, some recent studies revealed an overall induction of proteasome β-subunits (β5, β2, and β1) as an initial adaptive response to low-dose bortezomib exposure.20,21,38 Results from the present study indicate that in cells with established acquired resistance to bortezomib after chronic exposure to stepwise increasing doses of bortezomib, up-regulation of a mutant β5 protein serves as a compensatory mechanism to retain sufficient chymotrypsin-like activity. An enhanced expression of 20S proteasome core particle-associated β5 in drug-resistant cells (Figure 3C,D) may be consistent with this notion. Also data from proteasome activity labeling experiments (Figure 4C) indicate that at relatively low levels of bortezomib resistance (THP1/BTZ30) the bodipyFL-Ahx3L3VS probe showed attenuated binding/labeling to the mutant β5-subunit, but upon further up-regulation of mutated protein in THP1/BTZ50 and THP1/BTZ100 cells, labeling with the probe can increase concomitantly. It is not readily clear whether the up-regulation of constitutive β5 in bortezomib-resistant cells (Figure 3A,C,D) is accompanied by a concomitant decrease in immunoproteasome β5 expression as suggested by gene array data (Figure S2). This status may be normalized to control levels in the absence of bortezomib, as illustrated for THP1/BTZ(-100) cells (Figure 3A), but rapidly restored upon rechallenging with bortezomib (Figure 5C). The up-regulation of β5 was most pronounced in cells exposed to bortezomib concentrations up to 50 nM, a concentration range at which primarily β5-associated chymotrypsin-like proteasome activity, and to a lesser extent also β1-associated caspaselike activity, will be essentially abolished. Consistently, beyond concentrations of 50 nM bortezomib, induction of β5 expression is leveling off, as from this point inhibition of proteasome subunit β2-associated trypsinlike activity will be initiated.16,17
The present finding that the marked overexpression of β5 protein in bortezomib-resistant cells is not paralleled by induction of β5 mRNA levels points to a posttranscriptional regulatory mechanism. The nature of possible posttranscriptional effects in bortezomib-resistant cells is presently unclear but may possibly include alterations in proteasome homeostasis facilitated by an autoregulatory mechanism that mediates the differential polyubiquitination and degradation of multiple proteasome subunits, including β5.47 Hence, impaired polyubiquitination under bortezomib-selective pressure (Figure 1B) may differentially attenuate β5 degradation to yield a significant increase in β5 protein levels.
Under bortezomib-selective conditions, THP1/BTZ cells did not display any accumulation of ubiquitinated proteins (Figure 1B) or induction of stress-induced proteins (Figure S3), prototypical for the action of proteasome inhibitors. Beyond the effect of the mutant β5 protein, as well as its overexpression, it may be anticipated that the up to 2-fold elevated bortezomib-stimulated trypsin-like activity (Figure 4B) may compensate for the inhibition of chymotrypsin-like and caspase-like activities by sustaining proteasome activity above critical catalytic levels. Consequently, such a condition alone, or in combination with activities of deubiquitinating enzymes,48 for example, Poh149 as a possible candidate based on preliminary gene array studies (Figure S3), could also contribute to prevent the accumulation of toxic polyubiquitinated proteins, which may otherwise trigger loss of mitochondrial membrane potential and apoptosis.
We recently obtained evidence that a similar molecular basis of selective β5 induction after chronic exposure to bortezomib as described herein for THP1 cells also applies for bortezomib-resistant variants of human T-lymphocytic CCRF-CEM cells50 (Figure S5). Interestingly, when human 8226 myeloma cells were exposed according to the same strategy, only moderate (≤ 5-fold) resistance levels were observed.50 These results suggest that the onset of bortezomib resistance may differ significantly between hematologic tumor cell lineages. A possible lower propensity for multiple myeloma cells to acquire resistance to bortezomib may be consistent with the lack of detailed reports on this issue. Beyond this, in the case of multiple myeloma, host microenvironments may also play an important role in the efficacy of proteasome targeting.12 With the accumulating knowledge of potential mechanisms of resistance to bortezomib, more direct screening for these parameters should be the subject of future clinically directed laboratory studies. Screening for PSMB5 gene mutations in an isolated case of a bortezomib nonresponsive multiple myeloma patient did not provide evidence for mutations.51 However, based on the present observation that PSMB5 gene mutations may be provoked after exposure to clinically achievable concentrations of bortezomib in the range of 7 to 30 nM (Figure 6) screening for PSMB5 mutations should be reconsidered. In the context of proteasome activity and proteasome subunit composition, the recent identification and validation of specific proteasome-targeted probes38,52 may facilitate analyses on limited cell numbers.
It may be anticipated that the molecular mechanism of resistance reported in the current study specifically applies for bor-tezomib or other proteasome inhibitors that selectively target the β5-subunit of the proteasome. With the emergence of second-generation proteasome inhibitors targeting (ir)reversibly other proteasome subunits or multiple all β-subunits,13,19,53-55 it will be important to address whether they are able to circumvent bor-tezomib resistance, or that similar mechanisms of resistance are operative as described herein for bortezomib. If so, this would warrant the design of strategies to prevent selective up-regulation of specific proteasome subunits and thereby enhance the therapeutic efficacy of this novel class of therapeutic agents.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Prof M. H. Glickman (Department of Biology, Technion, Haifa, Israel), Dr H. Ovaa (Netherlands Cancer Institute, Amsterdam, The Netherlands), and Dr Frank Kruyt (Department of Medical Oncology, VU University Medical Center, Amsterdam, The Netherlands) are acknowledged for helpful discussions. Dr R. Honeywell (Department of Medical Oncology, VU University Medical Center) is acknowledged for excellent technical assistance.
This study is supported by a grant from the Dutch Arthritis Association (Amsterdam, The Netherlands; grant NRF-03-I-40 to G.J.), Cancer Center Amsterdam (Amsterdam, The Netherlands; STR Grant to J.C.), and ZonMW (The Netherlands Organization for Health Research and Development, The Hague, The Netherlands; N.E.F.).
Authorship
Contribution: R.O., N.E.F., Y.G.A., J.C., G.L.K., B.A.C.D., R.J.S., and G.J. designed research, analyzed and interpreted data, and wrote the paper; R.O., N.E.F., I.Z., G.L.S., K.D., C.L., J.W.H., C.R.B., G.J.P., and G.J. performed experimental research and analytical techniques; N.E.F., J.C., G.J., and B.Y. performed microarray analysis; K.V. and N.E.F. performed sequencing experiments; R.O., N.E.F., J.C., and G.J. collected data; and R.O. and G.J. performed statistical analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerrit Jansen, Department of Rheumatology, Rm 3A64, VU Institute for Cancer & Immunology, VU University Medical Center, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands; e-mail: g.jansen@vumc.nl.
References
Author notes
*R.O. and N.E.F. contributed equally to this work.