Telomeres are highly dynamic structures that adjust the cellular response to stress and growth stimulation based on previous cell divisions. This critical function is accomplished by progressive telomere shortening and DNA damage responses activated by chromosome ends without sufficient telomere repeats. Repair of critically short telomeres by telomerase or recombination is limited in most somatic cells, and apoptosis or cellular senescence is triggered when too many uncapped telomeres accumulate. The chance of the latter increases as the average telomere length decreases. The average telomere length is set and maintained in cells of the germ line that typically express high levels of telomerase. In somatic cells, the telomere length typically declines with age, posing a barrier to tumor growth but also contributing to loss of cells with age. Loss of (stem) cells via telomere attrition provides strong selection for abnormal cells in which malignant progression is facilitated by genome instability resulting from uncapped telomeres. The critical role of telomeres in cell proliferation and aging is illustrated in patients with 50% of normal telomerase levels resulting from a mutation in one of the telomerase genes. Here, the role of telomeres and telomerase in human biology is reviewed from a personal historical perspective.
Introduction
Since there are many excellent comprehensive reviews on the topic of telomeres, hematology, and aging,1,,–4 I decided that an autobiographic narrative might be more interesting and perhaps more entertaining. Of course, my personal story is just one of many. I understand that readers of Blood may not have time or patience for such trivia. Therefore, the first section, “Telomeres and hematology,” contains the most important scientific messages. The later sections provide a personal account of how my interest in stem cells led to studies of telomeres. My hope is that my personal and inevitably biased perspective will provide some insight in the development of these important research areas over the last 2 decades.
Telomeres and hematology
It is widely understood that telomeres represent the very ends of chromosomes, characterized by guanine-rich repetitive DNA and associated proteins.2 That telomerase is a reverse transcriptase that uses RNA to synthesize the G-rich repeats is also well known.1 The decrease in telomere length in most cells with proliferation and with age is appreciated, but what does it all mean? Despite intensive efforts by a large number of research teams, there is no definitive answer to this question. My hope is that, by breaking down the complex topic into smaller segments, parts of the answer will emerge.
Every chromosome end needs to be capped by a minimum number of telomere repeats to prevent activation of a DNA damage response.5,6 The simple way to think about this is that without a minimum number of telomere repeats any of the 92 ends of human chromosomes will resemble the end of a broken chromosome. Double-strand breaks are dangerous to cells and must be repaired prior to mitosis to prevent genome instability or cell death. So chromosomes must have a proper cap.
The length of the telomere repeat track determines the likelihood that a telomere will be properly capped. A minimum number of telomere repeats is required to recruit sufficient telomere binding proteins to fold the end into a properly capped structure. Most likely this structure consists of a T-loop in which the single-stranded 3′ end of the chromosome folds back into double-stranded telomere repeats.7 So to avoid activation of a DNA damage response, each telomere must have a minimum length of telomere repeats as well as fully functional “shelterin” proteins.2 Telomeric DNA together with various proteins form a proper cap.
The third point is that telomere length is maintained and set in the stem cells of the germ line that express high levels of telomerase. However, most somatic cells, including various stem cells, are unable to maintain telomere length. As a result, most somatic (stem) cells show progressive telomere shortening with each round of replication. B lymphocytes appear to be an exception to this rule in that memory B cells have longer telomeres than naive B cells.8 Elongation of telomeres in the B-cell lineage could reflect a requirement for extensive cell divisions imposed by clonal selection and affinity maturation of antibodies. Could it be that B cells are more likely to form tumors than T cells as a result?9 In general, the difference in telomere biology between the cells of the germ line and various somatic (stem) cells is poorly understood. One possibility is that telomeric chromatin is different, more “open” in the germ line (and perhaps the early embryo as well), increasing the chance of functional interactions between telomerase and chromosome ends. Other possibilities are that functional telomerase levels vary between cells of the germ line and somatic cells, for example, as a result of alternative splicing of hTERT transcripts10 or differences in the assembly or posttranslation modifications of the enzyme complex. Perhaps telomerase levels as well as telomere chromatin differ among various cell types.
The fourth point is that if a telomere becomes uncapped in a somatic cell, the outlook is not necessarily bleak as at least 2 telomere salvage pathways are available. Thus, following activation of a DNA damage response (via pathways that remain poorly understood), uncapped telomeres can be elongated by telomerase and/or recombination. Salvage by recombination requires the presence of homologous sequences and is probably less efficient when the average telomere length is short (ie, more efficient in mice than in humans11 ). Whether recombination or telomerase is used to salvage critically short telomeres varies between cell types and presumably depends on average telomere length, telomerase expression, and other factors. Telomerase appears to be the major salvage pathway for uncapped telomeres in hematopoietic cells. It is possible that homologous recombination reactions involving, for example, BRCA1 and BRCA2 and the Fanconi proteins are also important to sustain the proliferation of hematopoietic stem cells over a normal lifetime.
Unfortunately, there exists a third, dangerous pathway that extinguishes the DNA damage signals derived from uncapped telomeres: the fusions of 2 uncapped chromosome ends. These can be sister chromatids or the ends of 2 separate chromosomes. End-to-end fusions are dangerous as cells with intact cell cycle checkpoints will be fooled to enter mitosis with dicentric chromosomes. The anaphase bridges that are predicted to occur 50% of the time will typically induce chromosome breaks. The resulting genetic instability will kill most cells but loss or gain of genetic material can also provide a growth advantage to rare cells. Further fusions and mitotic break-fusion-bridge cycles can result in rampant genome instability. In this manner, telomere shortening is directly implicated in genome instability.
The sixth important point is that telomere salvage pathways have a limited capacity. Thus telomerase levels are limiting in most somatic cells. When more than a few uncapped telomeres accumulate in a cell (the exact number is not known and could differ among cell types as a function of the efficiency of the telomere salvage pathways), the capacity of the telomere salvage pathways is exceeded and cells either senesce or die by apoptosis. Both ATM and p53 are implicated in the DNA damage response originating from uncapped telomeres, and persistent DNA damage signals (eg, phosphorylated H2AX) originating from dysfunctional telomeres have been found in senescent cells.5 In healthy humans, this situation becomes increasingly likely for an increasing number of cells as their average telomere length decreases as a function of accumulated cell divisions and age. This is not necessarily a bad thing as telomere attrition provides a hurdle for aspiring tumor cells and thereby acts as a tumor suppressor mechanism. Indeed, it seems reasonable to assume that telomere length and telomerase levels evolved in humans to suppress the growth of tumors prior to reproduction and several decades of life. The flip side is that tight control over replication by telomere attrition probably results in accumulation of dysfunctional, senescent cells as well as significant loss of cells with age. The possibility that telomere attrition plays a more important role in human aging than in the aging of rodents and other model organisms does not sit well with many of my friends and colleagues who exclusively study model organisms. However, the striking clinical consequences of a 2-fold reduction in telomerase levels in humans (resulting from haploinsufficiency for one of the telomerase genes) in comparison with the absence of a phenotype for several generations in yeast, worms, plants, and rodents that completely lack telomerase strongly suggests that telomeres and telomerase play a role in controlling the proliferation of somatic cells in humans that does not exist as such in most model organisms including short-lived (inbred) mammals.
The final points relate to the pathological consequences of abnormalities in telomere maintenance. Mutations in genes encoding components of the telomerase enzyme including TERT, TERC, and DKC1 have been implicated in an increasing number of disorders including dyskeratosis congenita,4,12 aplastic anemia,13 and pulmonary fibrosis.14,15 Such mutations result in the assembly of dysfunctional telomerase complexes with partial activity or no catalytically activity whatsoever. Typically, only one allele of these telomerase genes is affected, resulting in a mixture of active and inactive telomerase enzyme complexes. There is little evidence to support dominant negative effects of inactive telomerase enzymes, and the net effect of mutations in genes encoding telomerase components is typically a reduction of up to 50% in overall telomerase levels.16 Dyskeratosis congenita is a bone marrow failure syndrome typically associated with skin pigmentation abnormalities, nail dystrophy, and leukoplakia. Other clinical manifestations are diverse in nature and can include lung fibrosis, liver cirrhosis, osteoporosis, and a predisposition to develop a variety of malignancies. It is important to note that telomerase deficiency per se does not cause pathology, but its consequences on telomere length do. These indirect effects can be separated into effects in the germ line (resulting in offspring with shorter telomeres) and somatic cells (resulting in a reduced efficiency of the telomerase-dependent telomere salvage pathway). The net result of the combined effects in the germ line and somatic cells is disease anticipation, the onset of disease occurring at earlier age in subsequent generations. The indirect relation between mutations in telomerase genes and the occurrence of a clinical phenotype has almost certainly resulted in an underestimation of the involvement of mutations in genes involved in telomere maintenance in various diseases by genetic linkage analysis. Most individuals with a mutation in one of the telomerase genes have no phenotype. Pathology occurs when cells are lost or impaired by uncapped telomeres to the point that the function of tissue or organ is compromised. The primary determinant of when such a threshold is reached appears to be the average telomere length.12 Cells at risks are cells that must sustain proliferation over a lifetime including lymphocytes and stem cells of tissues with a high turnover such the bone marrow. Factors in the environment or heritable factors that compromise the function or survival of (stem) cells in specific tissues increase the risk of telomere dysfunction in specific tissues. Such collaborative effects perhaps explain the diverse disease entities that have now been linked to telomerase deficiencies. Loss of (stem) cells in tissues, perhaps combined with changes in the marrow microenvironment,17 creates opportunities for abnormal cells that can respond to the growth stimulatory signals despite the presence of critically short and dysfunctional telomeres. The loss of telomere function, resulting from either telomerase disorders or exhaustion of stem cell following endogenous or exogenous damage, can thus result in aplastic and dysplastic changes as well as predispose for neoplastic growth. The intimate role of telomeres in aging18 and cancer19 assures that these fascinating dynamic structures will continue to be the subject of intense investigations over the next decades.
What to do?
One of my best childhood memories is crouching in the dunes behind my home in Wassenaar, the Netherlands, watching as gunpowder burned its way to a miniature rocket waiting in an improvised V-shaped rocket launching pad. After many failures, the rocket would fizz off into the sky and disappear out of sight. Ah, the sizzle, the sparks, the whoosh! This childhood experience has of course nothing to do with telomeres or hematology, but I was asked to provide a historic perspective. Sure enough I will get to stem cells and telomeres if I continue tracing my footsteps.
Like most teenagers, I did not have a clear picture of what to do with my life. I subscribe to the motto “when in doubt follow your nose but keep your options open.” The many different career options as an MD appealed to me. Nearing the end of my medical training, my main interest switched from psychiatry (having learned that sympathetic ears alone accomplish little) to internal medicine and immunology. At the time, it seemed reasonable to do “research,” although I was largely ignorant about the nature of such activities. After unproductive exercises in immunology, I decided to determine whether I could reproduce the work of Köhler and Milstein and make monoclonal antibodies.20 After many trials and errors, I eventually succeeded. How exciting it was to direct the growth, selection, and expansion of antibody-producing cells. In reproducing this published work, I had learned 2 important lessons: anything worthwhile can be reproduced and “practice makes perfect.” This was a turning point for me because it gave me the courage to choose a career in science.
My main focus initially was technical and, after further work on assays to screen for monoclonal antibodies against cell-surface antigens,21 hybridoma growth factor22,23 (which turned out to be IL-624,25 ), and the discovery of tetrameric antibody complexes,26 it became time for me to decide on a specific research area. I selected blood cell formation and hematopoietic stem cells, encouraged by the work of Stuart Schlossman and colleagues in Boston, who had shown that monoclonal antibodies are powerful tools to distinguish between morphologically similar cells in the immune system (Kung et al27 ). Inspired by the seminal work of Donald Metcalf,28 I believed the hematopoietic system was waiting to be “dissected” by monoclonal antibodies. By chance and the efforts of Allen Eaves, I ended up pursuing such studies in Vancouver, a truly spectacular place, secluded at the very border of the “civilized” world, yet a great place to pursue science, especially in the Terry Fox laboratory at the BC Cancer Research Center with colleagues such as Connie Eaves, Keith Humphries, and Gerry Krystal.
One of the first antibodies I produced in Vancouver was 8G12 specific for CD34.29 Curt Civin had shown that CD34 is a useful marker for stem and progenitor cells (Strauss et al30 ), and our antibody, also called HPCA-2, was found to be very useful to enumerate and study CD34+ cells. We also started exploring various applications of tetrameric antibody complexes,26 including applications for cell separation,31 which are now widely used.
Hematopoietic stem cells: where to start?
With “in-house” monoclonal antibodies and cell separation techniques at hand, we embarked in the late 1980s on activities that remain very popular to this day: purify stem cells as best as possible and study the behavior of such cells in culture in response to endless culture variables. The goal was to identify culture conditions that would allow meaningful “expansion” of stem cells in vitro for clinical applications. This strategy was based on compelling direct and indirect evidence supporting the “self-renewal” of stem cells in vivo. For example, in the mouse it is possible to isolate a single stem cell, transplant it into a lethally irradiated recipient, observe donor cell bone marrow reconstitution, and harvest many stem cells after a few weeks.32 Despite claims to the contrary,33 the best murine stem-cell purification procedures did not yet yield “pure” stem cells in the late 1980s, and my own work at the time was primarily focused on the human system. Apart from differences in telomere biology (discussed later) it is probably true that the frequency of stem cells in human bone marrow is several orders of magnitude lower than in the mouse. Jan Abkowitz and her collaborators in Seattle have made compelling arguments (Shepherd et al34,35 ) in support of the idea that the absolute number of stem cells and the number of times they divide are very similar in all mammals. If this is correct, the estimated 104 to 105 stem cells in the mouse are distributed over approximately 2 liters of bone marrow in humans. Based on this estimate, the frequency of stem cells in adult human bone marrow is expected to be less than 1 per 107 nucleated bone marrow cells. No wonder attempts to purify, genetically modify, or expand human adult stem cells have run into hard times. It seems very possible that the low frequency of actual stem cells in human bone marrow (and “mobilized” peripheral blood) represents a major, underappreciated challenge for both basic studies and clinical applications.
To avoid variables in the quality and quantity of human blood and bone marrow samples available for my research, I decided to work with previously frozen aliquots of bone marrow cells from cadaver organ donors. This was a great advance because it avoided biologic variation among samples and allowed systematic work on stem-cell purification, assays, and culture. Based on the work of Mike Dexter et al who had developed culture systems for murine hematopoietic cells,36 we developed a surrogate assay for human stem cells using limiting dilution strategies based on the their presumed ability to initiate long-term cultures.37 Other approaches to measure human stem cells were explored by John Dick in Toronto, who transplanted human stem cells into immune-deficient animals.38 Both assays have been very useful in human stem-cell research. However, doubts remain about the efficiency, reproducibility, and cell types measured in either assay relative to the cells that sustain normal hematopoiesis or the cells that repopulate bone marrow upon transplantation.
Another area of interest was the development of tissue culture medium for human hematopoietic cells.39 Most tissue culture media were developed and optimized for various immortal cell lines more than 50 years ago, and it seemed reasonable to assume that primary human hematopoietic cells could have different requirements. My work in this area was inspired by Norman Iscove, who had developed a superior serum-free medium for the growth of B cells and hybridomas (Iscove and Melchers40 ). In view of the doubts about which cells to study and the unknown growth factor requirements of primary hematopoietic (stem) cells, our progress was limited. Nevertheless, the recipe we developed (containing high levels of transferrin and low-density lipoproteins) is still used and probably provides a good starting point for further work in this important area.39,41
Developmental changes in stem-cell function?
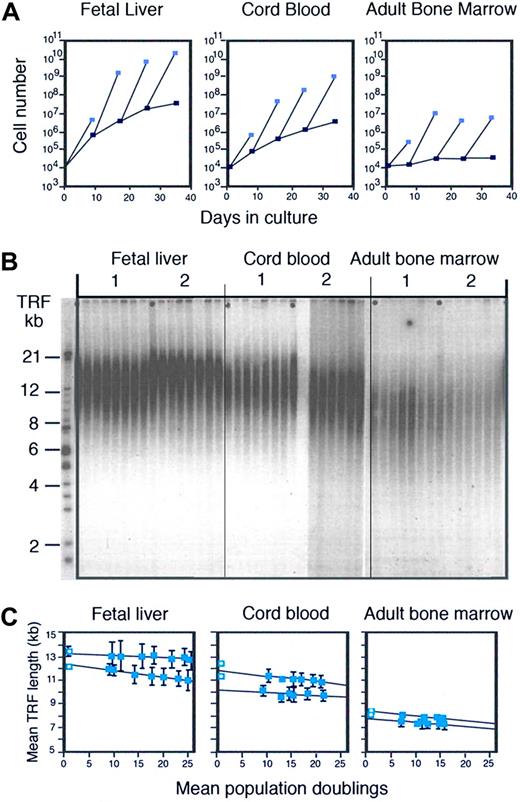
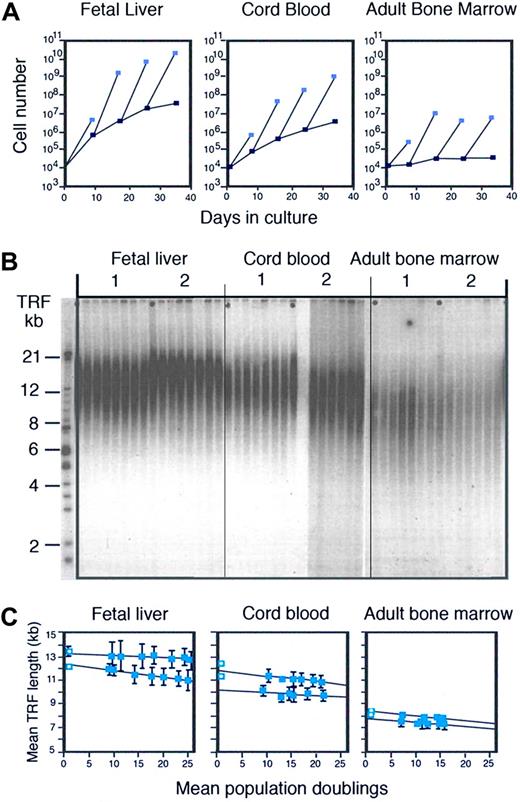
While the work on monoclonal antibodies, culture systems, tissue culture media, and cell separation techniques represented significant technical advances, it did not serve the main objective of my studies: to purify human stem cells and cultivate such cells to obtain a clinical meaningful numeric expansion in culture. On the contrary, the whole exercise seemed tedious and trivial when it became apparent that the maintenance of CD34+ cells in serum-free long-term cultures was the result of sequential recruitment of CD34+ cells that were initially quiescent.39 Frustrated by our inability to obtain any meaningful numeric expansion in culture with purified candidate stem cells from adult human bone marrow, we decided to explore the behavior of cells with the same phenotype purified from human cord blood and fetal liver. To our great surprise, the ability to produce CD34+ cells was hundreds to thousands of times better in cultures initiated with purified cells from cord blood and fetal liver (Figure 1A).42 Could the proliferation and expansion of stem cells be developmentally controlled? Because our findings greatly complicated simple notions about “stem cells” and “self-renewal,” they remain as unpopular today as they were 15 years ago. A sobering lesson for aspiring scientists: you can be right and make exciting advances but whether your work is accepted or acknowledged depends largely on social networks in science and is often independent of its quality or merit. The news is not all bad: most gems are eventually rediscovered. The good news for the investigator is that lack of recognition has advantages in terms of freedom to pursue novel research directions and time not wasted on making appearances.
Developmental changes and telomere loss in hematopoietic cells. (A) Ontogeny-related differences in the production of CD34+ cells in culture. (Reproduced from Lansdorp et al42 with permission.) Candidate stem cells with a CD34+CD45RAlowCD71low phenotype were purified from fetal liver (wk12), umbilical cord blood, and adult bone marrow and 104 sorted cells were cultured in serum-free culture medium supplemented with IL-6, IL-3, steel factor, and erythropoietin as described.39 At the indicated time interval, the total number of nucleated cells (light blue squares) and CD34+ cells (dark blue squares) present in the cultures was calculated from cell counts and the percentage of viable CD34+ cells measured by flow cytometry. All CD34+ cells from bone marrow cultures and fractions of the CD34+ cells from cord blood and fetal liver cultures were sorted and used for continuation of the cultures. (B) Loss of telomeric DNA in human hematopoietic cells upon proliferation in vivo and in vitro (reproduced from Vaziri et al43 with permission from the National Academy of Sciences of the United States). DNA samples from total nucleated cells from 2 different donors for each tissue before and after culture at increasing time points were subjected to terminal restriction fragment (TRF) size Southern blot analysis as described elsewhere.43 Cultures were initiated with highly enriched stem cells as described in panel A. (C) Quantitative analysis of the TRF data shown in panel B. The first time point (blue square with circle) represents the TRF value in nucleated cells from each tissue before purification. The mean and standard error of 2 independent TRF measurements (different gels) for each DNA sample purified from total nucleated cells at each time point are shown. The total number of cells was used to calculate population doublings. The loss of telomeric DNA in these cultures varied between 19 and 54 base pairs per population doubling. For details see Vaziri et al.43
Developmental changes and telomere loss in hematopoietic cells. (A) Ontogeny-related differences in the production of CD34+ cells in culture. (Reproduced from Lansdorp et al42 with permission.) Candidate stem cells with a CD34+CD45RAlowCD71low phenotype were purified from fetal liver (wk12), umbilical cord blood, and adult bone marrow and 104 sorted cells were cultured in serum-free culture medium supplemented with IL-6, IL-3, steel factor, and erythropoietin as described.39 At the indicated time interval, the total number of nucleated cells (light blue squares) and CD34+ cells (dark blue squares) present in the cultures was calculated from cell counts and the percentage of viable CD34+ cells measured by flow cytometry. All CD34+ cells from bone marrow cultures and fractions of the CD34+ cells from cord blood and fetal liver cultures were sorted and used for continuation of the cultures. (B) Loss of telomeric DNA in human hematopoietic cells upon proliferation in vivo and in vitro (reproduced from Vaziri et al43 with permission from the National Academy of Sciences of the United States). DNA samples from total nucleated cells from 2 different donors for each tissue before and after culture at increasing time points were subjected to terminal restriction fragment (TRF) size Southern blot analysis as described elsewhere.43 Cultures were initiated with highly enriched stem cells as described in panel A. (C) Quantitative analysis of the TRF data shown in panel B. The first time point (blue square with circle) represents the TRF value in nucleated cells from each tissue before purification. The mean and standard error of 2 independent TRF measurements (different gels) for each DNA sample purified from total nucleated cells at each time point are shown. The total number of cells was used to calculate population doublings. The loss of telomeric DNA in these cultures varied between 19 and 54 base pairs per population doubling. For details see Vaziri et al.43
Because the observed functional differences among hematopoietic cells at different stages of development were obtained using conditions that were identical, we proposed that the functional differences were intrinsic to cells at different stages of development.44 The very low frequency of actual stem cells in adult bone marrow mentioned above provides a caveat: it is possible that the cell populations purified from fetal liver and cord blood were far more enriched for actual stem cells than the candidate stem cells purified from human bone marrow. However, our data seemed to indicate that the behavior of stem cells could be primarily determined by unknown factors with a developmental component. If correct, dreams of stem-cell therapies starting with very few purified stem cells had run into a major roadblock. The idea of embarking on a quest to unravel the molecular mechanism that control stem-cell behavior and expansion at various stages of development furthermore did not appeal to me. I was aware of the limited progress in understanding the more accessible and better documented developmental clock regulating the expression of globin genes and the idea of similar studies with rare primary cells and unknown genes was unattractive. This was my mindset in early 1993 when I met Houmayoun Vaziri, who was interested in a position as a graduate student in my laboratory.
A mitotic clock in stem cells?
Vaziri asked me if I had looked at telomere length as a variable that could be related to the developmental changes in stem-cell function we observed. He showed me data on telomere length in human lymphocytes that he had just published together with Cal Harley and Richard Allsopp (Vaziri et al45 ). Looking at the smears of terminal restriction fragments (TRFs) obtained using Southern analysis, I remember being distinctly unimpressed. Nevertheless, the idea that fetal and adult stem cells could have differences in DNA that could be measured intrigued me and we decided to collaborate. One problem was that Southern analysis requires a lot of DNA (extracted from a million cells or more) and we could not purify that many human candidate stem cells. Instead, we decided to analyze the telomere length in the millions of cells that were readily produced in our serum-free cultures initiated with a few thousand purified candidate stem cells. The results of the blinded TRF analysis were shocking to me: the average telomere length in the cultured cells followed a very clear pattern: fetal liver longer than cord blood, and cord blood longer than adult bone marrow (Figure 1B). Furthermore, the telomere length showed a steady and progressive decline in culture (Figure 1C), in line with the idea that every cell division was accompanied by a loss of measurable number, estimated at 30 to 100 base pairs, of telomere repeats.43 To convince ourselves that our observations were not related to suboptimal culture conditions, we put our magnetic cell separation technique to work and we purified several million human CD34+ CD38− cells directly from more than 2 × 1010 organ donor bone marrow cells. The telomere length in those purified cells was longer than in CD34+ CD38+ cells but shorter than in the differentiated cells from fetal liver.43 Taken together, these results supported the idea that cell divisions are indeed “counted” in hematopoietic stem cells by progressive telomere loss. The immediate implication of this conclusion was that the self-renewal and proliferation of stem cells could eventually be limited by progressive telomere loss. It was time for me to learn more about telomeres.
Telomere length analysis by FISH?
In 1994, the telomere field was very small and spearheaded by a number of exceptional scientists including Elizabeth Blackburn, Ginger Zakian, Titia de Lange, and Carol Greider. In 1985, Carol Greider and Liz Blackburn had published their pioneering work on telomerase in the unicellular organism Tetrahymena,46 and a year later Howard Cooke, based on his finding that sperm DNA had longer telomeres than nucleated blood cells, suggested that telomerase levels in stem cells could be insufficient to maintain telomere length (Cooke and Smith47 ). Further support for this idea was obtained a few years later,48,49 and subsequent studies showed that the telomere length in fibroblasts predicted their replicative life span50 and that telomere shortening in immortal tumor cells is prevented because they express high levels of telomerase.51 For these studies, telomere length was invariably measured using TRF length analysis by Southern analysis. My first venture into the telomere field, after collaborating with Houmayoun Vaziri, was a quest to develop novel tools to measure telomere length at individual telomeres and in individual cells. This quest was driven by necessity. If telomeres were indeed important parameters in the biology of stem cells (I was now convinced of this), then better tools to study telomere length would be required. I was intrigued by the possibility of using fluorescence in situ hybridization (FISH) for such measurements, although I had no knowledge of FISH techniques. This was an advantage as several FISH experts whom I consulted invariably told me that to measure repetitive DNA with a length of less than 10 kb in a quantitative way using FISH was impossible. However, encouraged by Ger van den Engh and Michael Egholm, I was hoping that advances could be made using newly discovered peptide nucleic acid (PNA) probes,52 and I decided to give PNA a try during a sabbatical stay in the Netherlands with easy access to the outstanding “FISH” laboratory of Hans Tanke and Ton Raap at Leiden University. After many trials and errors, I finally managed to obtain reproducible results with human chromosome preparations. The key innovations in the technique that is now known as quantitative FISH (Q-FISH)53 were the use of high concentrations (70%) of formamide in the hybridization as well as the wash solutions, the use of low ionic strength to enable quantitative hybridization, and the inclusion of blocking protein to prevent nonspecific binding of the directly labeled, hydrophobic PNA probes specific for (TTAGGG)n repeats.
Chromosome-specific differences in telomere length
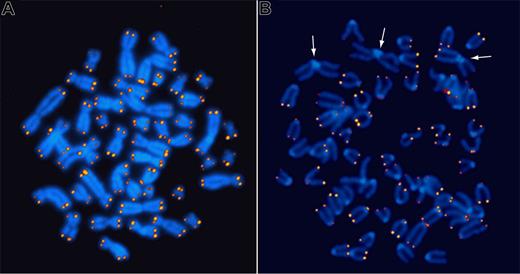
Q-FISH analysis of metaphase chromosomes from normal human cells revealed that every chromosome end carries a detectable number of telomere repeats and that the length of telomere repeats at individual chromosome ends is strikingly heterogeneous (Figure 2A). Immediate questions that arose were the following: How does the variation at individual ends relate to the progressive loss of telomere repeats in cells with each round of cell division? Are length differences chromosome specific? Could the presence of long telomeres support repair of short telomeres in the absence of telomerase? Does telomerase play a role in generating telomere length diversity? To start addressing some of these questions, I was keen to apply Q-FISH to cells from telomerase RNA (Terc) knockout (KO) mice that were rumored to have no phenotype whatsoever. When we obtained cells from such mice, we found that telomeres decreased in length by around 5 kb with each subsequent generation, and we readily observed chromosome fusions indicative of loss of telomere function in generation 6 (Figure 2B), the last (infertile) generation.54 Because this telomere length phenotype had been completely missed by conventional Southern analysis, the general interest in our Q-FISH technique greatly increased.
Fluorescence in situ hybridization to detect telomere repeats. Quantitative in situ hybridization (Q-FISH) using Cy3-labeled (CCCTAA)3 peptide nucleic acid probes on metaphase chromosomes from a human lymphocyte (A) and an embryonic fibroblast from a late-generation telomerase RNA KO mouse (B). Note that the fluorescence intensity is very similar for both sister chromatids at individual chromosome ends but very heterogeneous between ends. The arrows in panel B point to pseudo metacentric chromosomes resulting from the fusion of 2 acrocentric mouse chromosomes. Note the lack of telomere repeats at the junction of the 2 fused chromosomes.
Fluorescence in situ hybridization to detect telomere repeats. Quantitative in situ hybridization (Q-FISH) using Cy3-labeled (CCCTAA)3 peptide nucleic acid probes on metaphase chromosomes from a human lymphocyte (A) and an embryonic fibroblast from a late-generation telomerase RNA KO mouse (B). Note that the fluorescence intensity is very similar for both sister chromatids at individual chromosome ends but very heterogeneous between ends. The arrows in panel B point to pseudo metacentric chromosomes resulting from the fusion of 2 acrocentric mouse chromosomes. Note the lack of telomere repeats at the junction of the 2 fused chromosomes.
The telomere knockout mouse has been instrumental in elucidating the critical role of telomerase in telomere maintenance in mammals and has highlighted the critical role for telomeres in maintaining genome stability. The findings in this mouse model argue against a role of telomeres as a developmental clock: subsequent generations of mice with progressively shorter telomeres develop normally. However, the mouse model has proven to be somewhat misleading for human disease in that the consequences of partial telomerase deficiency in humans appear to be much more severe than that of complete lack of telomerase in mice. Most likely, the repair of critically short telomeres by homologous recombination in the absence of telomerase is more efficient when the average telomere length is long. Murine telomeres are on average approximately 10-fold longer than in humans.11
Q-FISH was then used to study the length of telomere repeats on specific human chromosomes.55 The striking heterogeneity in telomere length was not resolved by studies of specific chromosome ends. Interestingly, the average telomere length on chromosome 17p was found to be relatively short in all individuals tested, suggesting that this telomere will be one of the first to become uncapped upon progressive telomere shortening with proliferation and/or age in human cells. The rate of telomere attrition at the inactive X chromosomes in female cells was found to exceed that of the active X chromosome,56 indicating that differences in average length of specific chromosomes could be generated during proliferation and age. Differences in chromosome-specific telomere length were found to be partly heritable55,57 and partly originating stochastically in the germ line.58 Such differences could reflect differences in (sub)telomeric chromatin, repair by telomerase or recombination, timing of replication, or combinations of these factors.
Telomere length measurements by flow FISH
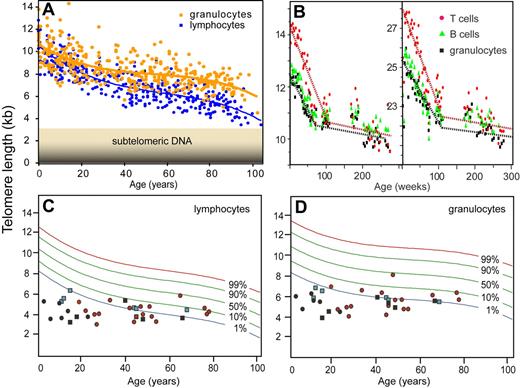
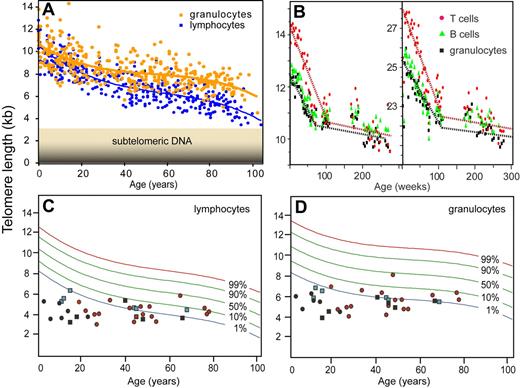
Q-FISH has been instrumental in the “telomere field” but the technique is very time-consuming and requires metaphase chromosomes that cannot always be obtained. So I was keen to explore alternative ways to measure telomere length using our PNA probes. In 1997, we started to explore in situ hybridization in suspension followed by measurements of specific fluorescence of individual cells using flow cytometry. Major challenges using this approach were keeping cells in suspension following denaturation of DNA required for hybridization and development of reproducible measurements while correcting for variable autofluorescence. Natalie Rufer and Tim Bruemmendorf in my laboratory worked to develop the initial technique,59,60 which was further improved by Gabriela Baerlocher et al.61 Key advances were the inclusion of bovine thymocytes in every tube as an internal reference and the use of a robotic device to increase the efficiency and reproducibility of the many wash steps in the protocol. Flow FISH has revealed that the decrease in telomere length with age is quite different for circulating granulocytes and lymphocytes and that, at any given age, telomere length shows a marked variation among individuals (Figure 3A). In collaboration with Neal Young, Rodrigo Calado, Hinh Ly, Blanche Alter, Sharon Savage, Mary Armanios, and others, we found that the “flow FISH” technique is very useful to screen for possible involvement of telomere defects in various disorders. Such disorders include diseases resulting from mutations in telomerase genes such as dyskeratosis congenita,12,63 aplastic anemia,64,65 and pulmonary fibrosis.14,15 In view of the highly significant decline in telomere length with age as well as the variation in average telomere length between individuals of the same age (Figure 3), it seems likely that these disorders represent only the tip of an iceberg and that other diseases with a “telomere component” will be discovered.
Telomere length measurements using flow FISH. (A) Nonlinear decline in telomere length with age. The average telomere length in granulocytes and lymphocytes from 392 healthy donors was calculated from the median telomere fluorescence and median autofluorescence relative to internal control cells (bovine thymocytes).61 Note that the telomere length at any given age is highly variable, that the most rapid drop in telomere length occurs early in life, and that the rate of telomere attrition in lymphocytes exceeds that in granulocytes. (B) Longitudinal studies of telomere length in newborn baboons point to a high turnover of hematopoietic stem cells in the first year of life.62 Note that the 2 animals differ markedly in average telomere length and that the rate of telomere attrition drops markedly after approximately 1 to 2 years in both animals. In humans, with a longer lifespan, this drop is expected to occur before the fourth year of life in line with previous cross-sectional observations.60 (C,D) Telomere length in individuals with mutations in telomere genes. The telomere length in cells from healthy individuals (A) was used to plot the telomere length distribution in the normal population using a best-fit approach (red, green, and blue curves representing expected telomere length for the indicated proportion of healthy individuals). The telomere length in lymphocytes (C) and granulocytes (D) from patients with known mutations in telomerase genes measured in the context of several studies12,–14,16,63 are shown. Each symbol represents an individual patient diagnosed with clinical symptoms associated with a mutation in dyskerin (DKC1, black circles), hTERT (red circles), and hTERC (black squares). Some individuals are carriers of a TERC mutation but have no clinical symptoms (blue square). The majority of individuals that carry mutations in telomerase genes display critically short telomeres, nearly all of them below the tenth percentile of the normal distribution and a majority of these below the first percentile (typically for both cell subsets shown). Note that individuals with early onset of disease (in the first 3 decades of life) show the most striking difference between observed and expected telomere length.
Telomere length measurements using flow FISH. (A) Nonlinear decline in telomere length with age. The average telomere length in granulocytes and lymphocytes from 392 healthy donors was calculated from the median telomere fluorescence and median autofluorescence relative to internal control cells (bovine thymocytes).61 Note that the telomere length at any given age is highly variable, that the most rapid drop in telomere length occurs early in life, and that the rate of telomere attrition in lymphocytes exceeds that in granulocytes. (B) Longitudinal studies of telomere length in newborn baboons point to a high turnover of hematopoietic stem cells in the first year of life.62 Note that the 2 animals differ markedly in average telomere length and that the rate of telomere attrition drops markedly after approximately 1 to 2 years in both animals. In humans, with a longer lifespan, this drop is expected to occur before the fourth year of life in line with previous cross-sectional observations.60 (C,D) Telomere length in individuals with mutations in telomere genes. The telomere length in cells from healthy individuals (A) was used to plot the telomere length distribution in the normal population using a best-fit approach (red, green, and blue curves representing expected telomere length for the indicated proportion of healthy individuals). The telomere length in lymphocytes (C) and granulocytes (D) from patients with known mutations in telomerase genes measured in the context of several studies12,–14,16,63 are shown. Each symbol represents an individual patient diagnosed with clinical symptoms associated with a mutation in dyskerin (DKC1, black circles), hTERT (red circles), and hTERC (black squares). Some individuals are carriers of a TERC mutation but have no clinical symptoms (blue square). The majority of individuals that carry mutations in telomerase genes display critically short telomeres, nearly all of them below the tenth percentile of the normal distribution and a majority of these below the first percentile (typically for both cell subsets shown). Note that individuals with early onset of disease (in the first 3 decades of life) show the most striking difference between observed and expected telomere length.
I have come to the end of my story on telomeres, stem cells, and hematology. A recurrent theme in my work has been that advances were enabled by the development and application of new techniques to address questions raised by unexpected observations. This is also how we more recently became interested in the genetic factors that regulate the stability of G-rich DNA66 and telomere length.67 The 2 closely related helicase genes identified in these studies are homologs of the human Fanconi J gene, and their precise role remains of great current interest. However, my latest “rocket launch” includes studies related to the possibility that sister chromatids differ in epigenetic marks at genes that regulate self-renewal and differentiation.68 As before, these studies require development of novel tools. I am very excited about this ongoing work and I feel extremely privileged to once again be in a position to experience the excitement that comes with exploring uncharted territory.
Acknowledgments
I specifically want to thank Rob Aalberse, Lucien Aarden, Janis Abkowitz, Blanche Alter, Sam Aparicio, Mary Armenios, Duncan Baird, Michael Barnett, Graham Betton, Rodrigo Calado, Chris Counter, Jeff Davis, John Dick, Roeland Dirks, Bob Donahue, Allen Eaves, Connie Eaves, Ger van den Engh, Fred Goldman, Carol Greider, Cal Harley, Lea Harrington, Phil Hieter, Richard Hodes, Keith Humphries, Al Klingelhutz, Gerry Krystal, Andras Nagy, Hihn Ly, Gordon Phillips, Steven Poon, Ton Raap, Dirk Roos, Ann Rose, Sharon Savage, Hergen Spits, Guy Sauvageau, John Schrader, Sara Selig, Salvatore Siena, Heather Sutherland, Hans Tanke, Leon Terstappen, Barb Trask, Houmayoun Vaziri, Neal Young, and Wim Zeijlemakers. I also want to thank the following current and former technicians and trainees in my laboratory for their contributions: Geraldine Aubert, Gabriela Baerlocher, Agnes Baross, Tim Bruemmendorf, Liz Chavez, Iris Cheung, Wieslawa Dragowska, Ester Falconer, Maloy Ghosh, Matt Greenwood, Prakash Hande, Mark Hills, Uwe Martens, Hector Mayani, Vivienne Rebel, Alex Roth, Nathalie Rufer, Mike Schertzer, Terry Thomas, Bert Wognum, Evert-Jan Uringa, Irma Vulto, and Mark Zijlmans. Janis Abkowitz, Gerry Krystal, Olga Lansdorp, and Claudia Bos are thanked for critically reading drafts of the paper. I apologize to all people whose work or contribution I forgot to mention.
Work in my laboratory is supported by grants from the National Institutes of Health (AI29524), the Canadian Institutes of Health Research (MOP38075 and GMH79042), and the National Cancer Institute of Canada (with support from the Terry Fox Run).
National Institutes of Health
Authorship
Conflict-of-interest disclosure: The author declares a financial interest in Repeat Diagnostic, a company specializing in leukocyte telomere length measurements using flow FISH.
Correspondence: Peter M. Lansdorp, Terry Fox Laboratory, BC Cancer Research Centre, 675 W 10th Ave, Vancouver, BC V5Z 1L3; e-mail: plansdor@bccrc.ca.