KIT is a tyrosine kinase receptor that is aberrantly activated in several neoplasms. In human pathologies, the most frequent mutation of KIT occurs at codon 816. The resulting KIT mutant protein is activated in the absence of ligand and is resistant to the clinically available inhibitors of KIT. In this report, we provide evidence for an essential function of the cytoplasmic tyrosine kinase FES downstream of KITD816V. FES is phosphorylated on tyrosine residues in cells that carry KITD816V mutation, and this phosphorylation is KIT dependent. Reduction of FES expression using RNA interference results in decreased cell proliferation in human or murine cells harboring KITD816V or the homologous mouse mutation KITD814Y. The reduced cell growth can be rescued using another cytokine (granulocyte-macrophage colony-stimulating factor [GM-CSF]) and is not observed when the closely related fer gene is targeted. Finally, signaling downstream of KITD816V is altered in cells lacking FES expression. This study shows a major function of FES downstream of activated KIT receptor and thereby points to FES as a novel target in KIT-related pathologies.
Introduction
The proto-oncogene c-kit encodes a receptor with tyrosine kinase activity which is essential for hematopoiesis and the development of melanoblasts, germ cells, and interstitial cells of Cajal. Gain-of-function mutations of KIT are implicated in human pathologies. These mutations have been classified as regulatory-type mutations when they affect domains of the receptor necessary for maintaining KIT autoinhibition and structural-type mutations when they directly affect the catalytic domain. Juxtamembrane mutations of KIT are commonly found in gastrointestinal stromal tumors (GISTs),1 whereas kinase domain mutations are mainly found in mastocytosis,2 hematologic neoplasms,3,–5 and germ-cell tumors.6,–8 Very recently, both juxtamembrane and kinase domain mutations of KIT have also been reported in melanomas.9 The most frequent mutation affects codon 816 in human c-kit cDNA, which corresponds to codon 814 in the homologous mouse sequence.10 Whereas tyrosine kinase inhibitors such as imatinib are very efficient on wild-type KIT (KITWT) and KIT juxtamembrane mutations, KITD816V is resistant to this treatment.11,–13 Therefore, drugs that could target this mutant or an essential downstream effector would be useful tools for the study and the treatment of the associated diseases.
Signaling proteins activated downstream of gain-of-function mutants of KIT include many proteins shown previously to participate in wild-type KIT receptor signal transduction.14,15 They include the classical phosphatidylinositol 3-kinase (PI3K) and extracellular signal-regulated kinase (ERK) pathways, as well as a number of signaling proteins activated by many cell-surface receptors such as p38, JNK, VAV, STAT-1, STAT-3, and STAT-5. Activation of PI3K is critical for both WT and mutant KIT in all models studied, while the activation of the mitogen-activated protein (MAP) kinases ERK-1 and ERK-2 appears to be dependent on the cellular context. The importance of each protein or signaling pathway activated downstream of KITD816V has yet to be fully evaluated.
Fps/fes was originally identified as an oncogene from avian (fps) and feline (fes) retroviruses. FES (feline sarcoma) and FER (FES-related) proteins are the only 2 members of a subfamily of cytoplasmic protein tyrosine kinase.16 FES has been shown to associate with growth factor and cytokine receptors,17,,–20 which leads to activation of its catalytic activity through tyrosine phosphorylation. The downstream effectors of FES are not clearly characterized, although several studies point to a link between FES and several signaling proteins such as STATs,21,–23 the MAP kinases ERK-1 and ERK-2,24,25 and PI3K.26 Several mouse models of FES have been generated with either a null allele,23,27 a knock-in of an inactive kinase, 22,28 a truncated FES protein,29 or transgenic mice expressing an active form of FES.25,30,31 They all showed a role for FES, albeit nonessential, in the homeostasis of the myeloid cell compartment, and a role in innate immunity has been highlighted in the knock-out mice.23,27,32 Mutations of FES have been recently reported in colorectal cancers.33 The mutant forms of FES appeared to have reduced kinase activity rather than being gain-of-function mutations.34,35 To date, the function of FES in normal physiology and in pathology remains enigmatic.34
This study was initiated to identify novel proteins required for KITD816V signaling. FES was found to interact with activated KIT receptor in yeast and was identified as a potentially phosphorylated protein in cells that express the KITD816V mutant. In human and mouse cell lines, FES is indeed phosphorylated downstream of KITD816V and downstream of the murine homolog KITD814Y, respectively. Using RNA interference to reduce FES protein expression, we show that FES is required for KITD816V-dependent cell proliferation both in cellular models with transfected KITD816V and in a mastocytoma cell line with endogenous expression of the mutant. In the absence of FES, the rate of proliferation is reduced due to slow progression of cells from G1 to S phase. Furthermore, cells treated with fes siRNA showed aberrant activation of STAT proteins and of mTOR pathway through p70 S6 kinase, suggesting a function for FES upstream of these signaling proteins.
Materials and methods
Yeast 2-hybrid screen
A yeast 2-hybrid screen was carried out as described previously.36,37 In brief, human c-kit cDNAs were expressed as the LexA fusion protein using the pBTM116 vector in the L40 yeast strain. A cDNA library cloned in pVP16 derived from the multipotential hematopoietic cell line EML-c1 (kindly provided by Dr S. Tsai, Seattle, WA) was screened using KITD816V as bait. The bait was functionally tested against the SH2 domains of p85 and growth factor receptor-bound protein 2 (GRB-2) before performing the screen.
Cells
TF-1 is a human erythroleukemia cell line, and TF-1 KITD816V cells are TF-1 cells stably transfected with the KITD816V receptor.38 P815 is a mouse mastocytoma cell line with an endogenous KITD814Y mutation. D816VKIT bone marrow-derived mast cells (BMMCs) are from transgenic mice expressing KITD816V.38 HMC-1 is a human mast cell line heterozygous for 2 mutations located on the same allele of c-kit: V560G and D816V (kindly provided by Dr J. H. Butterfield, Rochester, MN). All cells were maintained in RPMI 1640 medium, supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 100 U/mL penicillin-streptomycin (all from Invitrogen, Carlsbad, CA) and grown at 37°C in 5% CO2 at constant humidity. TF-1 cells were grown in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF; Berlex, Seattle, WA) or stem cell factor (SCF; Abcys, Paris, France).
siRNA transfection
Small interfering RNA (siRNA) and their sources were as follows. fes A siRNA was an equimolar mix of 2 RNA duplexes which target the human sequences 5′-GGCCAAGUUUCUACAGGAA-3′ and 5′-GCCUGAGGCUGAGUACCAA-3′ (Qiagen, Valencia, CA). The human fes B siRNA targeted the human c-fes sequence 5′-CCAACAUCGUGCGUCUCAUUGGUGU-3′ (Stealth siRNA; Invitrogen, Paisley, United Kingdom). The 2 human fer siRNAs were either a mix of the following 3 duplexes 5′-GGUGAAGUAUAUAAGGGCACAUUAA-3′, 5′-GCCCUAAGUUCAGUGAACUUCAGAA-3′, and 5′-ACGUAUCCAAGUCUUGGCUACUUAU-3′ (Stealth siRNA) or a mix of 4 siRNAs, SMARTpool M-003129-01-0005 (Dharmacon Research, Lafayette, CO). The human c-kit siRNA was as published39 (Qiagen). The mouse fes I siRNA targeted the sequence 5′-UGCGGCAGCAUGCAGAAGAUCUGAA-3′ (Stealth siRNA). The mouse fes II siRNA targeted sequence was 5′-GCAGAGAUAACAAGCCAGACCGAGA-3′ (Stealth siRNA). The control siRNA was 5′-UUCUCCGAACGUGUCACGU-3′ (Qiagen).
A total of 0.2 to 0.8 nmol siRNA were mixed with 5 to 15 × 106 cells in 0.5 mL RPMI 1640 medium in 4-mm electroporation cuvettes. Electroporation were done at room temperature using a Gene Pulser Electroporator II (Bio-Rad Laboratories, Munich, Germany) at 250 V and 400 μF for TF-1 cells40 and 300 V and 950 μF for P815 cells.
Antibodies
Anti-AKT, anti–phospho-AKT (Ser473), anti-KIT, anti-p38 MAP kinase, anti-mTOR, anti–phospho-mTOR (Ser2448), anti-p70 S6 kinase, anti–phospho-p70 S6 kinase (Thr389), anti–phospho-STAT1 (Tyr701), anti–phospho-STAT3 (Tyr705), and anti–phospho-STAT5 (Tyr694) rabbit polyclonal antibodies were from Cell Signaling Technology (Beverly, MA). Anti-ERK2, anti-KIT C-19, anti-STAT1, anti-STAT3, and anti-STAT5 rabbit polyclonal antibodies and anti-FES N-19 goat polyclonal antibody were from Santa Cruz Biotechnology (Heidelberg, Germany). Anti–active pan-ERK (Thr202/Tyr204) and anti–active p38 (Thr180/Tyr182) rabbit polyclonal antibodies were from Promega (Madison, WI). The other antibodies used were antiphosphotyrosine 4G10 (Upstate Biotechnology, Lake Placid, NY), anti-FES F113 rat monoclonal antibody (Calbiochem, Darmstadt, Germany), anti-FER rabbit polyclonal antibody (Millipore, Bedford, MA), anti–rat RARa/IgM rabbit polyclonal antiserum (Nordic Immunology, Tilburg, the Netherlands), mouse IgG2b (Transduction Laboratories, Lexington, KY), and rat IgM (Zymed, South San Francisco, CA).
Immunoprecipitation and Western blotting
Cells washed in phosphate-buffered saline (PBS) were lysed in HNTG buffer (50 mM HEPES [pH 7], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2, and 1 mM EGTA) containing protease inhibitor mixture (Roche Applied Science, Mannheim, Germany), 50 mM NaF, and 100 μM Na3VO4. Protein concentration was measured using the Bio-Rad Protein Assay Kit (Bio-Rad Laboratories). For immunoprecipitation, the samples were incubated with antibody and a bed volume of 10 μL protein A- or protein G-sepharose (Amersham Biosciences, Uppsala, Sweden), washed 3 times with HNTG buffer, and eluted in Laemmli buffer at 100°C for 5 minutes. Proteins were separated on SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membrane (Immobilon-P; Millipore). Membranes were incubated with antibodies and treated using Supersignal West pico chemiluminescent substrate (Pierce, Rockford, IL).
Immunoprecipitations of human FES were done with a mix of F113 antibody and anti-RARa/IgM. Phosphotyrosine immunoprecipitations were done with the 4G10 antibody covalently linked to agarose beads (Upstate Biotechnology).
Cell proliferation assay
A total of 104 cells/well were seeded into 96-well plates in 100 μL of RPMI 1640 medium with 10% fetal bovine serum with or without 5 ng/mL GM-CSF or 250 ng/mL human SCF (Abcys). Cells were incubated for 24 hours at 37°C and pulsed for 6 hours with 0.0185 MBq (0.5 μCi) of [methyl3H]-thymidine (Amersham Biosciences). Cells were then transferred onto glass fiber filters (Packard Instruments, Groningen, the Netherlands), and incorporation was measured using a Rackbeta Compact 1212-411 β-counter (LKB, Uppsala, Sweden).
For carboxyfluorescein succinimidyl ester (CSFE) labeling, 5 × 106 cells were washed with PBS and labeled with 5 μM CFSE (Molecular Probes, Leiden, the Netherlands) for 10 minutes at 37°C. Cells were then washed 2 times with culture medium. Fluorescence of cells was analyzed by flow cytometry. A total of 2 × 104 events was collected on a FACScan (Becton Dickinson, San Jose, CA) and analyzed using FlowJo software (Ashland, OR).
BrdU incorporation assay
106 cells were incubated for 15 minutes at 37°C with 1 mM BrdU solution. BrdU and 7-amino-actinomycin D (7-AAD) staining was performed according to the BrdU Flow kit manual (Becton Dickinson, San Diego, CA). A total of 4 × 104 events was collected on a FACScan and the cellular DNA content was analyzed by FlowJo software.
Results
Identification of novel effectors of KIT signaling
To identify new intracellular proteins in the oncogenic pathway of the KIT receptor, 2 strategies were undertaken. A yeast 2-hybrid screen was performed using KITD816V intracellular domain fused to LexA DNA-binding domain as bait and a cDNA library from EML-c1 cells. One of the isolated clones interacted with KITD816V but not with the control baits LexA-Lamin and KITW42, a loss-of-function mutant of KIT. This clone corresponded to FES tyrosine kinase cDNA. In parallel, mass spectrometry was used to identify tyrosine-phosphorylated proteins in cells that express KITD816V, and FES was again identified by this method.
FES is phosphorylated downstream of human KITD816V and murine KITD814Y
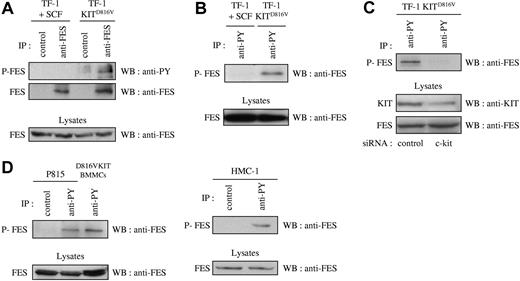
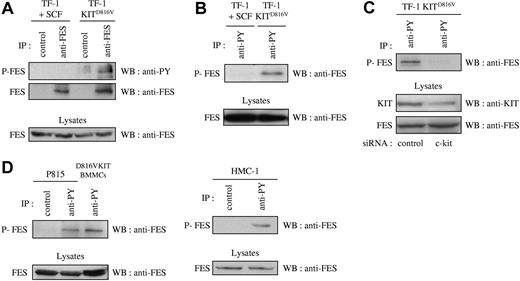
Previous published work has correlated the phosphorylation status of FES with its activation status.41,–43 If FES is a downstream effector of KIT signaling, it is expected to be activated and therefore phosphorylated on tyrosine residues. To analyze the phosphorylation status of FES in cells that express the D816V mutant, we first used the cell-line TF-1 KITD816V and the parental TF-1 cells that express endogenous KIT protein. Endogenous FES protein expression has been reported in this human erythroleukemic cell line.17 The parental cells can be propagated either with GM-CSF or with KIT-ligand SCF, while TF-1 KITD816V cells proliferate in the absence of exogenous cytokines. Phosphorylation of FES downstream of KITD816V was analyzed by immunoprecipitation of FES followed by Western blotting with an antiphosphotyrosine antibody. Whereas no phosphorylation of FES on tyrosine residues was detected in TF-1 cells grown in SCF, FES was phosphorylated in TF-1 KITD816V (Figure 1A). Accordingly, FES was detected following immunoprecipitation of phosphorylated proteins with the antiphosphotyrosine 4G10 antibody in TF-1 KITD816V, but not in the parental cells (Figure 1B). To demonstrate the direct implication of KITD816V in FES activation, KIT expression was reduced using siRNA, and FES phosphorylation was analyzed. As shown Figure 1C, reduced expression of KIT resulted in dramatic decrease of FES phosphorylation.
FES is phosphorylated in cells that harbor KITD816V. Immunoprecipitations (IPs) were carried out on soluble cell lysates (SCLs) of serum-starved cells. SCLs were also directly probed with anti-FES to show equivalent quantity of proteins in all lysates. (A) IPs were done on SCLs of TF-1 cells with anti-FES antibody or with an isotype-matched irrelevant antibody (IP control), followed by Western blotting (WB) with an antiphosphotyrosine antibody (anti-PY) or anti-FES antibody. (B) IPs were done on SCLs from TF-1 cells with anti-PY antibody, followed by WB with anti-FES antibody. (C) Lysates from TF-1 KITD816V cells treated with siRNA were immunoprecipitated using anti-PY antibody, followed by WB with anti-FES antibody. KIT and FES protein expression were also controlled in the cell lysates. (D) IPs on lysates from mouse (P815 and D816VKIT BMMCs) or human (HMC-1) mastocytomas with anti-PY antibody or with an isotype-matched irrelevant antibody, followed by WB with anti-FES antibody. P-FES indicates phosphorylated FES protein. The data presented in each panel are representative of 1 out of at least 3 independent experiments.
FES is phosphorylated in cells that harbor KITD816V. Immunoprecipitations (IPs) were carried out on soluble cell lysates (SCLs) of serum-starved cells. SCLs were also directly probed with anti-FES to show equivalent quantity of proteins in all lysates. (A) IPs were done on SCLs of TF-1 cells with anti-FES antibody or with an isotype-matched irrelevant antibody (IP control), followed by Western blotting (WB) with an antiphosphotyrosine antibody (anti-PY) or anti-FES antibody. (B) IPs were done on SCLs from TF-1 cells with anti-PY antibody, followed by WB with anti-FES antibody. (C) Lysates from TF-1 KITD816V cells treated with siRNA were immunoprecipitated using anti-PY antibody, followed by WB with anti-FES antibody. KIT and FES protein expression were also controlled in the cell lysates. (D) IPs on lysates from mouse (P815 and D816VKIT BMMCs) or human (HMC-1) mastocytomas with anti-PY antibody or with an isotype-matched irrelevant antibody, followed by WB with anti-FES antibody. P-FES indicates phosphorylated FES protein. The data presented in each panel are representative of 1 out of at least 3 independent experiments.
We then investigated FES phosphorylation in mastocyte cell lines. HMC-1 is a human mast-cell leukemia cell line with 2 point mutations at codons 560 and 816 in the c-kit gene. P815 and D816VKIT BMMCs are 2 models of murine mastocytes that carry the homologous murine D814Y mutation endogenously and human KITD816V, respectively. All 3 were analyzed for phosphorylation status of FES. Again, FES was present in the phosphotyrosine immunoprecipitates in all 3 mastocyte cell lines (Figure 1D). These results show constitutive phosphorylation of FES downstream of KITD816V/D814Y mutants.
FES but not FER mediates proliferation of TF-1 KITD816V and P815 cells
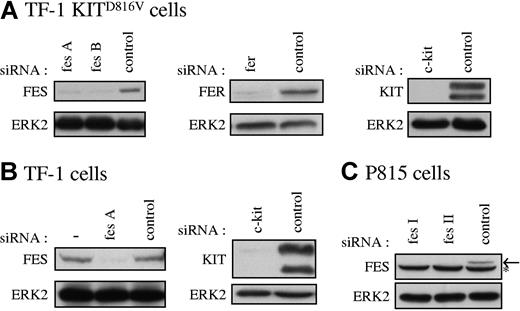
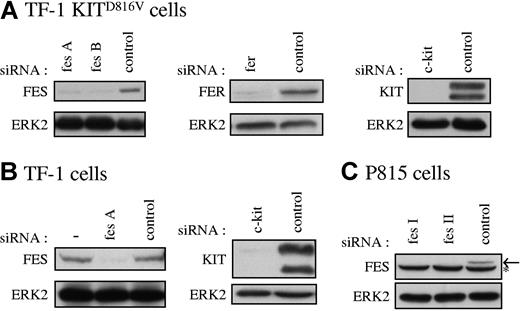
Our data suggested that FES tyrosine kinase is activated downstream of KITD816V and not downstream of the wild-type KIT receptor. To determine if FES is implicated in KITD816V-mediated cell growth, we used RNA interference to abolish FES protein expression. As controls in our experiments, we also used siRNAs directed against c-kit and against the fes-related gene fer. Figure 2 illustrates the reduction of expression of FES, FER, and KIT achieved in representative experiments following transfection of the cell lines with siRNAs. All siRNA experiments described were systematically controlled by Western blotting.
Specific reduction of FES, FER, and KIT proteins in cultured cells. FES, FER, and KIT protein expression in TF-1 KITD816V cells (A), TF-1 cells (B), and P815 cells (C) treated with the corresponding specific siRNAs were analyzed by Western blot. SCLs were prepared 48 hours following transfection, and 30 μg of proteins were loaded per lane. As loading control, ERK2 expression was assessed. fes A and fes B are 2 independent human fes siRNAs. fes I and fes II are 2 independent mouse fes siRNAs (“siRNA transfection”). (B) — indicates lysate of cells not treated with siRNA. (C) Arrow shows FES protein. *Nonspecific band in P815 cells.
Specific reduction of FES, FER, and KIT proteins in cultured cells. FES, FER, and KIT protein expression in TF-1 KITD816V cells (A), TF-1 cells (B), and P815 cells (C) treated with the corresponding specific siRNAs were analyzed by Western blot. SCLs were prepared 48 hours following transfection, and 30 μg of proteins were loaded per lane. As loading control, ERK2 expression was assessed. fes A and fes B are 2 independent human fes siRNAs. fes I and fes II are 2 independent mouse fes siRNAs (“siRNA transfection”). (B) — indicates lysate of cells not treated with siRNA. (C) Arrow shows FES protein. *Nonspecific band in P815 cells.
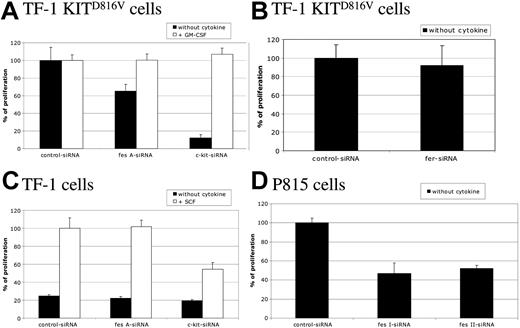
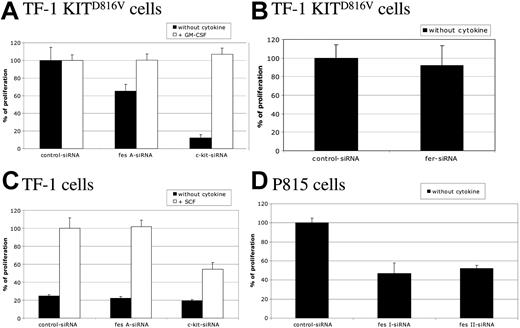
To study the biological function of FES downstream of KIT, cell proliferation of TF-1 KITD816V cells was first assayed by [methyl3H]-thymidine incorporation. These cells strictly depend on KIT for cell proliferation, as shown by the cell sample treated with c-kit siRNA (Figure 3A). Silencing of the fes gene in TF-1 KITD816V resulted in a decreased cell proliferation of 35% when compared with control siRNA. A similar reduction was also observed using another independent fes siRNA (data not shown). Furthermore, addition of GM-CSF to the culture medium, which complements the requirement for KIT signaling (Figure 3A; c-kit siRNA + GM-CSF), rescued the cell-proliferation defect associated to FES inactivation (Figure 3A; fes siRNA), indicating specificity of FES in KITD816V-mediated cell proliferation. In these experiments, inactivation of fer did not affect TF-1 KITD816V cell proliferation (Figure 3B).
FES is required for KITD816V-induced cell proliferation. Following siRNA treatment, cells were seeded in 96-well plates and maintained with or without cytokine as indicated. Thymidine incorporation assays were done 24 hours later. Cells were TF-1 KITD816V (A,B), TF-1 (C), and P815 (D). Cell proliferation is represented as a percentage of control siRNA. The data are from 3 independent experiments done in triplicate. Error bars indicate the standard error of the mean.
FES is required for KITD816V-induced cell proliferation. Following siRNA treatment, cells were seeded in 96-well plates and maintained with or without cytokine as indicated. Thymidine incorporation assays were done 24 hours later. Cells were TF-1 KITD816V (A,B), TF-1 (C), and P815 (D). Cell proliferation is represented as a percentage of control siRNA. The data are from 3 independent experiments done in triplicate. Error bars indicate the standard error of the mean.
We also investigated the proliferation of the parental TF-1 cells in response to SCF (Figure 3C). These cells do not proliferate in the absence of exogenous cytokines (Figure 3C; black histograms). In the presence of SCF, c-kit siRNA, but not fes siRNA, reduced the proliferation of TF-1 cells, indicating that FES is not required for KITWT-mediated cell proliferation.
Finally, we addressed whether FES is required for cell proliferation in mastocytomas using the P815 cell line, which expresses KITD814Y mutant endogenously (Figure 3D). Proliferation and survival of P815 cells is strictly dependent on KIT, as the use of either the KIT inhibitor dasatinib (BMS-354825) or the reduction of KIT expression using siRNA resulted in cell death within 24 hours (data not shown). We abolished FES expression using 2 independent murine siRNAs. As observed in TF-1 KITD816V, fes silencing resulted in decreased proliferation of P815 cells by 50% (Figure 3D). These results indicate that FES is required for cellular proliferation downstream of the kinase domain mutant KITD816V/KITD814Y.
FES facilitates G1 to S phase transition
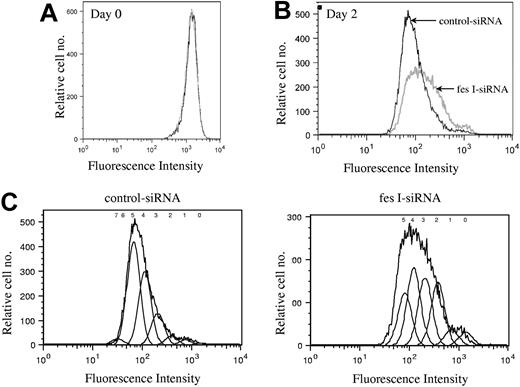
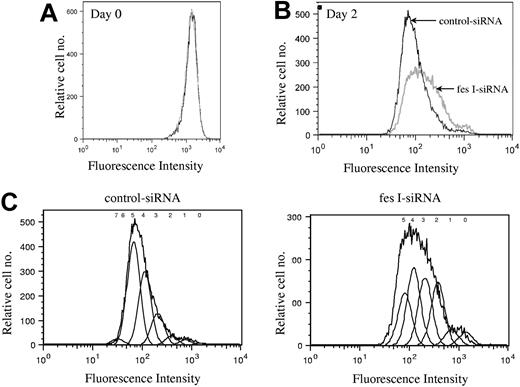
We next investigated whether FES is required for cell survival or for cell-cycle progression. Annexin V and 7-AAD staining did not show any evidence for cell death in the absence of FES (data not shown). To delineate the role of FES in cellular proliferation, we stained the cells with CFSE and evaluated the number of cell divisions at various time points following staining. CFSE irreversibly couples to cellular proteins; when cells divide, the fluorescence is then equally distributed in the daughter cells. Cells transfected either with fes siRNAs or control siRNA presented identical profiles following staining (Figure 4A). The staining intensity was then used to evaluate the number of cell divisions in the 2 transfected populations. On day 2, the profile of the control siRNA-treated population was homogenous (Figure 4B,C), with most cells having done 5 divisions. In comparison, the FES-depleted population presented a wider peak with stronger labeling intensity, reflecting the fact that many cells had gone through 2, 3, or 4 cell divisions only (Figure 4B,C). Therefore, the decreased [methyl3H]-thymidine incorporation in the absence of FES is due to reduced rate of cell division rather than to cell death. We concluded that fes gene silencing resulted in slower proliferation of the cells.
Depletion of FES disturbs cell cycle. P815 cells transfected with either control or fes siRNAs were labeled with CFSE. Histogram plots represent the profiles for the 2 populations, fes I siRNA (gray line) and control siRNA (black line) on day 0 (A) and 2 days following labeling (B,C). (C) The estimated number of cell divisions is indicated above the histogram. The analyses were done with FlowJo software. These profiles represent 1 of 3 independent experiments that gave similar results.
Depletion of FES disturbs cell cycle. P815 cells transfected with either control or fes siRNAs were labeled with CFSE. Histogram plots represent the profiles for the 2 populations, fes I siRNA (gray line) and control siRNA (black line) on day 0 (A) and 2 days following labeling (B,C). (C) The estimated number of cell divisions is indicated above the histogram. The analyses were done with FlowJo software. These profiles represent 1 of 3 independent experiments that gave similar results.
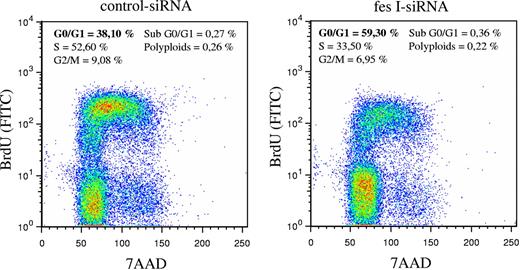
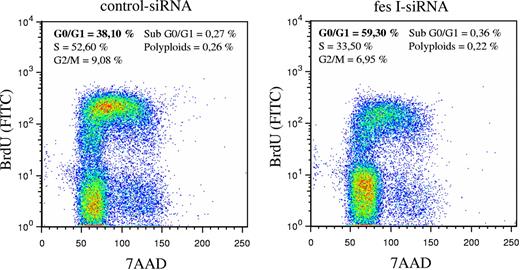
In parallel to these experiments, we analyzed the cell cycle using BrdU incorporation and DNA staining with 7-AAD. BrdU staining of cells was done 48 hours following electroporation of siRNAs. Whereas in our experimental conditions, most cells transfected with control siRNA entered in S phase (52% in S phase and 38% in G0/G1 phase), fes siRNA-treated cells were mainly in G0/G1 phase (59% versus 33% in S phase; Figure 5). Thus, reduced expression of FES resulted in accumulation of cells in G0/G1, suggesting that FES facilitates the transition between G1 and S phase. Altogether, these results demonstrate a function of FES on cellular proliferation downstream of the KIT kinase mutant.
Absence of FES slows down transition from G1 to S phase. P815 cells transfected with either control or fes siRNAs were stained with anti-BrdU-FITC antibody and 7-AAD. The percentage of cells in G0/G1, S, or G2/M phases were quantified using FlowJo software. The values are from 1 of 3 independent experiments that gave similar results.
Absence of FES slows down transition from G1 to S phase. P815 cells transfected with either control or fes siRNAs were stained with anti-BrdU-FITC antibody and 7-AAD. The percentage of cells in G0/G1, S, or G2/M phases were quantified using FlowJo software. The values are from 1 of 3 independent experiments that gave similar results.
FES acts negatively on STATs and positively on p70 S6 kinase activation pathway
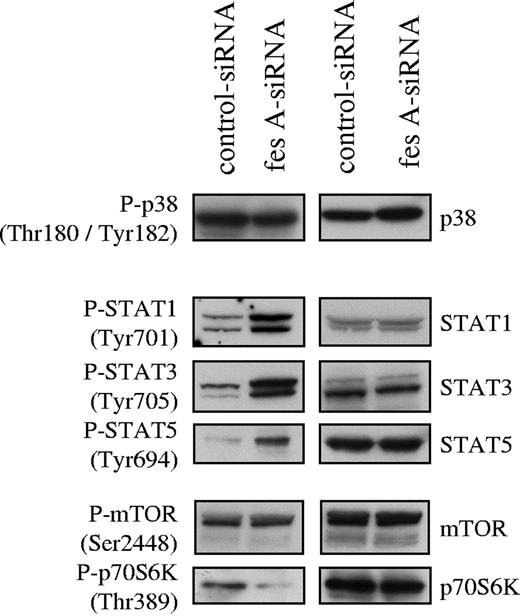
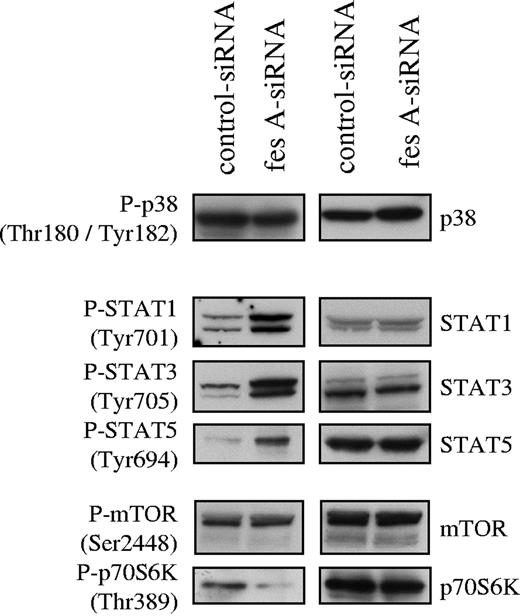
To investigate FES contribution to KITD816V downstream signaling, the activation status of several mediators of KIT signaling was analyzed. Cell lysates from control and fes siRNA TF-1 KITD816V cells were probed with phosphospecific antibodies. First, we analyzed the activation of the MAP kinases p38 (Figure 6) and ERK1/2 (data not shown). Their activation was not affected by down-regulation of FES expression. Subsequently, we looked at tyrosine phosphorylation of STATs. We found constitutive tyrosine phosphorylation of STAT-1, STAT-3, and STAT-5 in the control cells as previously described for KITD816V. The depletion of FES led to increased phosphorylation of all 3 STAT proteins (Figure 6). We also analyzed downstream effectors of the PI3K cascade. AKT (data not shown) and mTOR (Figure 6) phosphorylation were not modified following FES inactivation. By contrast, phosphorylation of Thr389, which is required for activation of p70 S6 kinase,44,–46 was decreased in cells lacking FES protein (Figure 6). These results suggest that FES negatively regulates the activation of STATs, but acts positively on p70 S6 kinase activation downstream of the activated KIT mutant.
Effect of fes siRNA on phosphorylation of STAT proteins and p70 S6 kinase. TF-1 KITD816V cells treated with either control or fes siRNAs were serum-starved for 16 hours. SCLs were immunoblotted with indicated anti-phospho antibodies and after stripping, reprobed with specific antisera. Similar results were obtained in 3 independent experiments.
Effect of fes siRNA on phosphorylation of STAT proteins and p70 S6 kinase. TF-1 KITD816V cells treated with either control or fes siRNAs were serum-starved for 16 hours. SCLs were immunoblotted with indicated anti-phospho antibodies and after stripping, reprobed with specific antisera. Similar results were obtained in 3 independent experiments.
Discussion
FES, FER, and cell-surface receptors
As an intracellular tyrosine kinase, FES is a downstream effector of signals transduced from cell-surface receptors, such as IL-3, GM-CSF, IL-4 receptors, and gp130-containing receptors.17,,–20,47 In primary erythroid cells, the activation of FES and/or FER downstream of wild-type KIT receptor has been suggested using a dual-specific antibody.25 Another study showed increased FER phosphorylation in response to SCF in BMMCs.48 The function of FER in KIT-mediated adhesion and chemotaxis was suggested through the use of a dominant-negative mutant protein.48 The function of these proteins in the context of gain-of-function KIT mutants has not been explored previously. In our models, RNA interference was used to significantly reduce gene expression of FES and FER. We show that FES was dispensable for cell proliferation mediated by KITWT, but is required for the KITD816V proliferation signal. This observation highlights differential signaling mechanisms between wild-type and mutant KIT receptors. In mouse models that harbor kinase-inactivating mutations in both genes, the redundant function of FES and FER has been suggested.16,28 Interestingly, our results indicate that FER does not compensate or participate in KITD816V-mediated proliferation.
FES regulates STAT proteins and p70 S6 kinase
In TF-1 KITD816V cells, FES regulates p70 S6 kinase and STAT-1, STAT-3, and STAT-5 activation, but not ERK1/2, p38, AKT, or mTOR. Interestingly, a similar function of FES upstream of p70 S6 kinase without affecting AKT phosphorylation has already been established in IL-4 signaling in the B lymphoma cell line M12.4.1.20 Several lines of evidence have documented a role for FES in regulating STAT tyrosine phosphorylation. Ectopic expression of FES increased tyrosine phosphorylation of STAT-3 and STAT-5,21 while monocytes from transgenic mice with a kinase-inactive FES show reduced STAT-3 and STAT-5 phosphorylation upon GM-CSF stimulation.22 However, consistent with our observations using RNA interference, in FES knock-out mice, STAT-3 and STAT-5 tyrosine phosphorylation is increased downstream of IL-6 and GM-CSF.23 A working model to reconcile all these observations is that STAT proteins are sequestered and phosphorylated by FES.23 Therefore, our observations and conclusions regarding FES function in KITD816V signaling are consistent with hypotheses raised in other systems. Whether the aberrant STAT tyrosine phosphorylation and/or the impaired activation of p70 S6 kinase are responsible for the proliferative defects remains to be elucidated. A function for p70 S6 kinase in cell growth is consistent with a number of previous studies on this kinase.49,–51
Signaling downstream of KITD816V
KIT gain-of-function mutations activate a large spectrum of proteins. The determination of which proteins are actually required for the oncogenic properties of KIT mutants is an important challenge. Some studies have addressed the requirement for some of these proteins for KIT function. Thus, PI3K is essential for survival and subsequently for KITD816V- or KITD814Y-mediated proliferation.52 In Mo7e cells, STAT-3 but not STAT-1 seems to be required for cell proliferation driven by KITD816V, as suggested by the use of dominant-negative mutants of these proteins.53 Using chemical inhibitors, it has been suggested that NF-κB, mTOR, and PKCδ are important effectors of KITD816V.54,–56 These hypotheses have not yet been challenged using gene inactivation techniques. Using antisense oligonucleotides and RNA interference, another study very recently implicated the BCL-2-related protein MCL-1 in KITD816V-dependent cell proliferation.57 We have shown here that FES is a component of KITD816V downstream signaling. Furthermore, using several independent siRNAs, we demonstrated that FES is required for cell proliferation downstream of KITD816V. However, fes siRNA reduced (35% and 50% reduction of TF-1 KITD816V and P815 cells respectively) but did not abolish cell proliferation (Figure 3A,D). Therefore, FES is 1 of several components required for cell proliferation. Mutations at codon 816 are the most frequent mutations of KIT. Yet, other KIT gain-of-function mutations occur in about 70% of GISTs, mainly affecting the regulatory juxtamembrane region. It remains to be determined whether FES is also necessary for the function of juxtamembrane mutants of KIT. Moreover, because of structural and signaling similarities among receptors, our study raises the question of the implication of FES downstream of other constitutively activated tyrosine kinase receptors.
Inhibitors of KITD816V
Several recent studies have identified inhibitors that reduce the growth or the survival mediated by KITD816V in cell cultures. These include drugs that target KIT itself such as dasatinib,58 AMN107,59,60 PP1 and PP2,61 semaxinib (SU5416),62 and PKC412,60,63,64 and drugs that target NF-κB,54 mTOR,56 protein kinase C δ (PKCδ),55 PI3K,52 and heat shock protein 90 (HSP90).65 The present study points to FES as a putative target of KITD816V-mediated cell proliferation. In contrast to NF-κB, mTOR, and PI3K, FES has limited nonredundant functions in normal physiology. Since FES is a kinase, selective ATP competitive inhibitors can be developed. The use of inhibitors for signaling components downstream of an oncoprotein are being considered to target the PI3K pathway.66,67 Considering that KIT kinase mutants show resistance to clinically available KIT inhibitors, and that secondary resistance occur in some patients, this strategy could be used either as an alternative to, or in combination with, the KIT inhibitors.
In conclusion, this study highlights the function of FES in cell proliferation downstream of the kinase-mutant KIT receptor. Since FES is dispensable for wild-type KIT signaling, it is an attractive putative therapeutic target in KIT-related pathologies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jean-Rémy Galindo and Sandrine Sarrazin for help with FACS analysis, J. H. Butterfield for the HMC-1 cell line, and Sheela Ramanathan for critical reading of the manuscript.
This work was supported by INSERM, La Ligue Contre le Cancer “équipe labellisée” and a grant from le Ministère de la Recherche (ACI [Action Concertée Incitative] Molécules et Cibles Thérapeutiques). E.V. was supported by a PhD fellowship from INSERM-Région PACA [Provence-Alpes-Cote d'Azur], and S.L. by a postdoctoral fellowship from la Fondation de France. P.D. and P.D.S. are INSERM scientists.
Authorship
Contribution: E.V. performed the experiments, analyzed and interpreted the data, and drafted the manuscript. S.L. contributed vital reagents. P.D. interpreted the data, approved the data, and provided logistic and financial support. P.D.S. contributed by designing and supervising the study, writing the manuscript, and approving the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paulo De Sepulveda, INSERM UMR599, 27 Blvd Leï Roure, 13009 Marseille, France; e-mail:sepulveda@marseille.inserm.fr.