Avet-Loiseau and colleagues present the definitive study of the clinical importance of genetic subtypes of multiple myeloma (MM).
These aberrations, detected by interphase fluorescence in situ hybridization (FISH), define subtypes of multiple myeloma (MM), each with different biology and prognosis and needing tailored management approaches. Avet-Loiseau and colleagues confirm the clinical relevance of genetic categories,1 using the largest clinical data set reported to date: a cohort that is uniformly treated and has sufficient follow-up time. Two genetic categories deserve special mention: t(4;14)(p16;q32) and 17p13 deletions. The former, a primary genetic aberration in MM, is observed in 15% of cases and is consistently associated with an unfavorable outcome (irrespective of treatment modality).1 Patients with t(4;14)(p16;q32) have short disease control with high-dose chemotherapy,2 and urgently need development of alternative therapeutic strategies, perhaps bortezomib in combination with alkylators.3 Patients with 17p13 deletions (P53 locus), a genetic progression event in MM, represent the highest risk of all MM genetic categories.1 These patients have a short survival and lower likelihood of response, and frequently will develop extramedullary disease, hypercalcemia, and plasmacytomas.1,4 A minor limitation of the study by Avet-Loiseau et al (because of constraints of available material for testing) is that it lacks the data on patients with t(14;16)(q32;q23). This translocation, alongside with the t(4;14)(p16;q32), is associated with aggressive MM.1 Other less common genetic variants have been described including CCND2, CCND3, and other MAF translocations. These subtypes are rare and thus lack reliable data on their clinical phenotype, response rate, and so forth (even when extrapolations and predictions are tantalizing when using gene expression profiling) (see figure).
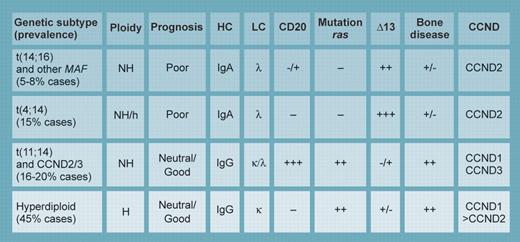
The most common and best defined subtypes of myeloma. NH indicates nonhyperdiploid; H, hyperdiploid; HC, preferred immunoglobulin heavy chain used; LC, preferred immunoglobulin light chain used by this genetic subtype; CD20, likelihood of CD20 expression in the cell surface; mutation ras, likelihood of ras mutation in this genetic subtype; deletion 13, likelihood of having chromosome 13 monosomy or deletion by genetic subtype; bone disease, prevalence by genetic subtype; and CCND, type of cyclin D gene up-regulated (by cis or trans mechanism) by the genetic subtype. Illustration by Kenneth Probst.
The most common and best defined subtypes of myeloma. NH indicates nonhyperdiploid; H, hyperdiploid; HC, preferred immunoglobulin heavy chain used; LC, preferred immunoglobulin light chain used by this genetic subtype; CD20, likelihood of CD20 expression in the cell surface; mutation ras, likelihood of ras mutation in this genetic subtype; deletion 13, likelihood of having chromosome 13 monosomy or deletion by genetic subtype; bone disease, prevalence by genetic subtype; and CCND, type of cyclin D gene up-regulated (by cis or trans mechanism) by the genetic subtype. Illustration by Kenneth Probst.
A novel observation is that β2-microglobulin can further stratify patients with high-risk genetic features—something in need of validation in independent data sets. Many factors beyond biology influence β2-microglobulin serum concentration, and will confound its incorporation into future prognostic models. An elevated β2-microglobulin concentration does not necessarily indicate a more aggressive MM subtype, but rather a more afflicted patient. Thus, an elevated β2-microglobulin concentration may not, in and of itself, dictate a change to a different therapy strategy (perhaps even force dose reductions!), in stark contrast to genetic factors. Accordingly, it is uncertain whether β2-microglobulin will have a place in the future of prognostic factors for MM. Of interest, the t(4;14)(p16;q32) and −17p13 could further subdivide all stages of the recently published International Staging System for MM.5
The obvious consequence of this study, and other similar ones in MM, is that we can no longer ignore these important clinical parameters, as they have a profound influence on the proper management of patients. For instance, we must not rely solely on high-dose chemotherapy as primary treatment modality for t(4;14)(p16;q32) or 17p13 MM. These data are now validated by several series and are not experimental; arguably, it is a disservice to patients not to include these data in patient evaluation. Because of the ease of using FISH as a clinical test, we have adopted the profiling of patients for a panel of genetic aberrations as part of our standard of standard of care. And all of this without using CHIPs (yet …)!
The author received consulting fees (less than $10 000) from Millennium Pharmaceuticals. ▪