Paroxysmal nocturnal hemoglobinuria (PNH) is characterized by the presence in the patient's hematopoietic system of a large cell population with a mutation in the X-linked PIG-A gene. Although this abnormal cell population is often found to be monoclonal, it is not unusual that 2 or even several PIG-A mutant clones coexist in the same patient. Therefore, it has been suggested that the PIG-A gene may be hypermutable in PNH. By a method we have recently developed for measuring the intrinsic rate of somatic mutations (μ) in humans, in which PIG-A itself is used as a sentinel gene, we have found that in 5 patients with PNH, μ ranged from 1.24 × 10–7 to 11.2 × 10–7, against a normal range of 2.4 × 10–7 to 29.6 × 10–7 mutations per cell division. We conclude that genetic instability of the PIG-A gene is not a factor in the pathogenesis of PNH.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is characterized by the presence of a large population of hematopoietic cells with an acquired somatic mutation in the X-linked PIG-A gene.1 PIG-A encodes an enzyme subunit involved in the biosynthesis of glycosylphosphatidylinositol (GPI), which is essential for the expression on the cell surface of the complement inhibitors CD55 and CD59 as well as a host of other proteins.2,3 The GPI– phenotype is the hallmark of the PIG-A mutant population, which is referred to as the PNH clone, which typically coexists with normal stem cells in patients at a variable frequency, typically from 10% to over 90%.4 However, we found that very small PIG-A mutant clones also exist in healthy individuals without ever undergoing clonal expansion,5 and this has recently been confirmed by others.6 The existence of PIG-A mutant clones in healthy individuals supports the immune escape model, where immunologic marrow injury selects for these clones in patients with the disorder but not in healthy individuals.7-9 This model is also supported by several observations: (1) GPI– cells have no intrinsic growth advantage in animal models10 ; (2) aplastic anemia and PNH frequently occur together11 ; (3) normal stem cells are lost12 and oligoclonal T-cell populations are expanded in PNH,13 as they are in aplastic anemia; and (4) HLA-DR 15 and 16 (DR-2) alleles are overrepresented among PNH patients as in aplastic anemia.14
Several different PIG-A mutant clones have been found in some patients with PNH.15-18 Indeed, it is not uncommon for patients to harbor distinct red cell populations with a partial and complete lack of CD59 (termed PNH II and PNH III cells4 ), which likely reflects the presence of distinct clones with partial and complete loss of GPI-anchor synthesis, respectively.17 While the PIG-A mutant clones in patients are at least 4 orders of magnitude larger than in healthy individuals (eg, 1:5 versus 1:100 000),5 smaller relative increases in the frequency (f) of lymphocytes with mutations in HPRT (Xq26-q27.2)19-21 and red cells with variants in glycophorin A (4q28.2-q31.1)22 have also been found in some patients with PNH. These findings have been regarded as suggestive of an increased mutation rate in PIG-A itself.15,19,20,22,23 To investigate this hypothesis, we analyzed μ in PNH using the PIG-A gene itself as a sentinel gene.24 The 5 patients whom we studied are representative of patients in whom the hypermutability hypothesis would be most applicable: those with large PNH granulocyte populations and evidence of oligoclonality based on the presence of both PNH II and PNH III red cells.4 In all cases, we found that the mutation rate was in the normal range.
Study design
Patients with “classic PNH” (Table 1) referred to Memorial Sloan-Kettering Cancer Center were recruited to the study and provided blood samples with informed consent on a protocol approved by the Institutional Review Board. Immortalized B-lymphoblastoid cell lines (BLCLs) were generated as previously described.26 Typically, BLCL populations from PNH patients either are entirely GPI+ or have both GPI+ and GPI– subpopulations. We were interested in the GPI+ cells from PNH patients, because they would provide a phenotypic marker for spontaneous PIG-A mutations: a change from the GPI+ to a GPI– phenotype. From 5 PNH patients we could generate GPI+ BLCLs (Table 2). The mutation rate in the PIG-A gene was determined as recently described.24 This method takes advantage of flow sorting to eliminate preexisting mutants from the BLCL population, expansion of the collected GPI+ cells in vitro, and analysis of a large number of cells to identify rare GPI– cells that have arisen due to spontaneous mutations in PIG-A in vitro.
Results and discussion
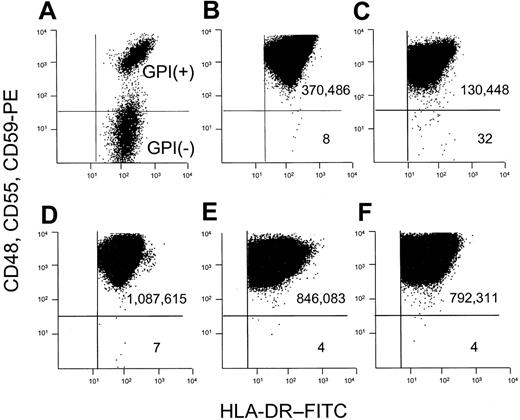
We used a combination of several antibodies that bind to distinct GPI-linked proteins. GPI– cells in a mixture with GPI+ cells are thus readily identified (Figure 1A). Occult GPI– cells arising spontaneously within a BLCL from a healthy donor are also clearly distinguished from the much larger GPI+ population from which they arise (Figure 1B). Using a cell line from a patient with ataxia-telangiectasia as a positive control for hypermutability, we observed an increased frequency of spontaneously arising GPI– cells (Figure 1C).
We used this combination of anti-CD48, anti-CD55, and anti-CD59 antibodies to analyze cell populations that had been expanded in vitro after preexisting mutants were purged by flow sorting (Figure 1D-F). This step allowed us to measure the mutation rate by the formula μ = f ÷ d.24 The mean calculated mutation rate for 5 patients with PNH was 4.6 × 10–7 mutations per cell division (Table 2), and all 5 were well within the normal range that we have reported (2.4 × 10–7 to 29.6 × 10–7 mutations per cell division).24
The presence of an expanded GPI– population and the finding of several different simultaneous15,17,18,27 or successive16 clones with distinct PIG-A mutations had been regarded as indicative of an increased mutation rate. The increased frequency of circulating HPRT mutants and glycophorin A variants in PNH had also been taken as evidence of hypermutability.19,20,22,23 Chen et al21 replicated such findings but provided data implicating increased cell divisions rather than hypermutability as the explanation. Our work now addresses the issue directly in 2 ways: (1) We use the same gene that is mutated in PNH as our sentinel gene, and (2) we eliminate preexisting mutants from the population to control for cell divisions and to calculate the mutation rate. From our findings, we conclude that an increased rate of mutation in PIG-A is not a part of the pathogenesis of PNH.
Our finding of a normal mutation rate in PNH is consistent with the observation that patients with this disorder are not particularly prone to cancer,11 whereas patients with inherited genomic instability (eg, ataxia-telangiectasia and Fanconi anemia) are not prone to PNH. Thus, hypermutability of the PIG-A gene is neither necessary nor sufficient for the development of PNH. While an increased number of cell divisions could result in an increased frequency of mutants,28 this must not be sufficient to result in PNH, because PNH is not seen after recovery from chemotherapy-induced marrow injury. Therefore, we conclude that the expansion of PIG-A mutant clones occurs as a result of selection in favor of the mutants, which occurs only in certain forms of marrow injury, such as in aplastic anemia.7-9 Having ruled out hypermutability in PNH, we predict that the direct measurement of the mutation rate will be useful in elucidating the pathophysiology of other clonal hematologic disorders.
Flow cytometric dot plot analysis for the calculation of f and μ. BLCLs were analyzed after staining sequentially with a mixture of unconjugated anti–human CD48, CD55, and CD59, PE-conjugated rabbit anti–mouse immunoglobulin antibody, and anti-HLA-DR–FITC. The number of events is shown in each quadrant. The mutant frequency (f) is calculated as the number of GPI– cells divided by the number of GPI+ cells. (A) Normal GPI+ cells mixed with GPI– cells from a patient with PNH. (B) BLCLs from healthy donor 1, where the frequency of mutants is 22 × 10–6 (C) BLCLs from a patient with ataxia-telangiectasia, with increased GPI– cells (f = 245 × 10–6). (D-F) To calculate the mutation rate, preexisting mutants are eliminated from the population by collecting the upper 50th percentile of the distribution curve after staining with anti-CD59. The collected GPI+ cells are then returned to culture and expanded. The mutation rate is determined by the formula μ = f ÷ d, where d represents the number of cell divisions occurring in vitro after sorting.24 (D) Analysis of a BLCL from healthy donor 2 after expansion after flow sorting. f = 6.4 × 10–6, d = 8 cell divisions, and μ = 8 × 10–7 mutations per cell division. (E-F) Analyses of 2 representative cell lines from PNH patients. (E) Analysis of a BLCL from PNH patient 1 after expansion after flow sorting. f = 4.73 × 10–6, d = 4.24 cell divisions, and μ = 11.2 × 10–7 mutations per cell division. (F) Analysis of a BLCL from PNH patient 2 after expansion after flow sorting. f = 5.05 × 10–6, d = 10.6 cell divisions, and μ = 4.76 × 10–7 mutations per cell division.
Flow cytometric dot plot analysis for the calculation of f and μ. BLCLs were analyzed after staining sequentially with a mixture of unconjugated anti–human CD48, CD55, and CD59, PE-conjugated rabbit anti–mouse immunoglobulin antibody, and anti-HLA-DR–FITC. The number of events is shown in each quadrant. The mutant frequency (f) is calculated as the number of GPI– cells divided by the number of GPI+ cells. (A) Normal GPI+ cells mixed with GPI– cells from a patient with PNH. (B) BLCLs from healthy donor 1, where the frequency of mutants is 22 × 10–6 (C) BLCLs from a patient with ataxia-telangiectasia, with increased GPI– cells (f = 245 × 10–6). (D-F) To calculate the mutation rate, preexisting mutants are eliminated from the population by collecting the upper 50th percentile of the distribution curve after staining with anti-CD59. The collected GPI+ cells are then returned to culture and expanded. The mutation rate is determined by the formula μ = f ÷ d, where d represents the number of cell divisions occurring in vitro after sorting.24 (D) Analysis of a BLCL from healthy donor 2 after expansion after flow sorting. f = 6.4 × 10–6, d = 8 cell divisions, and μ = 8 × 10–7 mutations per cell division. (E-F) Analyses of 2 representative cell lines from PNH patients. (E) Analysis of a BLCL from PNH patient 1 after expansion after flow sorting. f = 4.73 × 10–6, d = 4.24 cell divisions, and μ = 11.2 × 10–7 mutations per cell division. (F) Analysis of a BLCL from PNH patient 2 after expansion after flow sorting. f = 5.05 × 10–6, d = 10.6 cell divisions, and μ = 4.76 × 10–7 mutations per cell division.
Prepublished online as Blood First Edition Paper, March 16, 2006; DOI 10.1182/blood-2006-01-0256.
Supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award, HL56778-06, and NCI CA-109258.
Presented in part at the American Society for Hematology Annual Meeting, Philadelphia, PA, December 8, 2002.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.