The ZNF198/FGFR1 fusion kinase associated with an atypical myeloproliferative disease is constitutively activated and regulates several STAT transcription factors. We used oligonucleotide microarrays to compare the gene-expression profiles between HEK-293 cells that stably express either the ZNF198/FGFR1 chimeric protein or the wild-type ZNF198 gene. Expression of the plasminogen activator inhibitor-2 (PAI-2/SERPINB2) was highly increased in cells expressing the fusion gene. Western blot analysis demonstrated that HEK-293 cells do not express PAI-2 endogenously, but in ZNF198/FGFR1-expressing cells 2 molecular forms of PAI-2, which were 47 kDa and 32 kDa, were expressed intracellularly, and a 60-kDa form was secreted. Similarly, expression of ZNF198/FGFR1 in BaF/3 mouse hematopoietic cells also induced the expression of the PAI-2 protein. Immunoprecipitation analysis revealed that both intracellular forms of PAI-2 bind to the ZNF198/FGFR1 kinase. Treatment of HEK-293 and BaF/3 cells with TNF-α in the presence of cycloheximide, induced apoptosis in both cases. In contrast, HEK-293 and BaF/3 cells expressing ZNF198/FGFR1 were resistant to TNF-α-induced apoptosis. These observations suggest that expression of the ZNF198/FGFR1 fusion gene is associated with specific PAI-2-mediated resistance to apoptosis which may contribute to the highly malignant nature of leukemic cells carrying this fusion kinase gene.
Introduction
Reciprocal chromosomal translocations in hematologic malignancies often result in the generation of novel fusion proteins. These chimeric proteins generally provide novel signaling functions that are considered to account for the oncogenic events in the leukemic cells.1 One such chromosomal translocation was described in patients with an atypical form of myeloproliferative disease (MPD) that is associated with T-cell leukemia/lymphoma and peripheral-blood eosinophilia2 and involves a reciprocal t(8;13)(p11;q12) rearrangement.3-8 In this translocation a zinc finger gene, ZNF198, is fused in-frame with the fibroblast growth factor receptor 1 (FGFR1). The resultant protein contains the zinc finger motif and proline-rich domain of ZNF198 and the intracellular tyrosine kinase domain of FGFR1.9-12 The resulting ZNF198/FGFR1 fusion protein is a ligand-independent, constitutively active cytoplasmic tyrosine kinase, unlike FGFR1, which is a membrane-bound receptor tyrosine kinase. Although the signaling functions of the ZNF198/FGFR1 fusion kinase were proposed based on the functions of FGFR1, the fusion kinase has been suggested to have unique signaling functions that are different from FGFR1 because of its cytoplasmic localization and constitutive tyrosine kinase activity.13-14 The oncogenic potential of the ZNF198/FGFR1 fusion kinase was demonstrated in IL-3-dependent Ba/F3 cells, where stable expression of the chimeric fusion protein produced IL-3-independent growth.15-17 Although activation of FGFR1 is known to result from dimerization following ligand binding, the ZNF198/FGFR1 fusion kinase is activated through homodimerization of the ZNF198 zinc finger domain.14-16
ZNF198 is a widely expressed gene that encodes a 1377-amino acid nuclear protein with a molecular mass of 155 kDa.9-12 Prominent features of ZNF198 are the 5 zinc finger motifs and a proline-rich domain (PRD) within the central part of the protein and an acidic domain at the C-terminal end of the protein.9 On the basis of the structure and the type of zinc fingers present in ZNF198, it has been predicted to have a function in protein-protein interactions. A putative nuclear localization signal (NLS) is located in the C-terminal end of the ZNF198 protein, which is lost in the ZNF198/FGFR1 fusion protein. It is thought that the predominantly cytoplasmic localization of ZNF198/FGFR1 results from loss of the NLS.
Although it is generally assumed that the ZNF198/FGFR1 protein is responsible for the transformed phenotype in MPD, it is also possible that the cytoplasmic localization of the fusion protein may act in a dominant-negative way on the ZNF198 protein, because the 2 proteins can dimerize through the zinc finger motif.14 Apart from its dimerization role in the fusion gene, however, little is known about the normal function of ZNF198. We reported that ZNF198 interacts with the HHR6A/6B and RAD18 proteins that are involved in the postreplication DNA repair process.18 The ZNF198/FGFR1 fusion protein, however, only interacted with the HHR6A/6B proteins and not with HRAD18, which is essential for the DNA repair function. As a result, cells expressing the ZNF198/FGFR1 chimeric protein showed a reduced capacity to repair UV radiation-induced DNA damage.18
ZNF198/FGFR1-mediated signaling mimics that of ligand-activated FGFR1, in that both activate the signal transduction proteins STAT1, STAT3, and STAT5.14 The activation of STAT proteins in the presence of ZNF198/FGFR1 has also been observed in different cell types such as BaF3, HepG2, Cos7, and HEK-293,11,14,19 demonstrating that ZNF198/FGFR1 mediates the activation of these proteins irrespective of the cell type used. In the case of ZNF198/FGFR1 signaling, the activation of the STAT proteins was shown to be more potent and ligand independent. It has also been demonstrated that the ZNF198/FGFR1 protein shows the signaling functions comparable to those of the interleukin-6 cytokine receptors.14 To investigate potential dysregulation of gene activity in cells expressing the ZNF198/FGFR1 fusion protein, we generated several HEK-293 cell clones with constitutive activation of the fusion gene.14,18 RNA from these cells was used to screen the Affymetrix U133A oligonucleotide array. A consistent finding between different clones was the highly up-regulated expression of the plasminogen activator inhibitor-2 (PAI-2/SERPINB2) gene compared with the parental HEK-293 cells which did not show any detectable PAI-2 expression. The same up-regulation of PAI-2 was also seen in ZNF198/FGFR1-expressing BaF/3 hematopoietic cells. In both cases the cells are resistant to TNF-α/CHX-mediated cytotoxicity, only when the PAI-2 gene is activated.
Materials and methods
Constructs
The chimeric ZNF198/FGFR1 fusion gene was amplified by polymerase chain reaction (PCR) from leukemic cells expressing the fusion gene. The PCR product was then cloned into the pEGFP-C2 vector as an N-terminal GFP fusion gene. The full-length ZNF198 gene was amplified from a fetal bone marrow cDNA library and similarly cloned into the pEGFP-C2 vector. The full-length FGFR1 gene was amplified from HEK-293 cDNA and cloned into the pCDNA3 vector.
Cell culture and transfection studies
HEK-293 cells were maintained in DMEM with 10% FBS in 5% CO2 and were used to establish cells stably expressing the ZNF198 or the ZNF198/FGFR1 fusion kinase. Transfections were performed using lipofectamine (Invitrogen, Carlsbad, CA) as described earlier.18 Cells were transferred 48 hours after transfection into DMEM containing 10% FBS and 500 μg/mL G418. Green colonies were identified using a fluorescence microscope and isolated using ring cloning. BaF/3 cells were routinely maintained in RPMI-1640 containing 10% FBS (complete medium) with 1 ng/mL IL-3. Transfection into these cells was achieved by electroporation. Briefly, 5 × 106 cells were washed 3 times in 1 × PBS, mixed with 20 μg linearized ZNF198/FGFR1 plasmid, and electroporated using a Bio-Rad (Hercules, CA) gene pulser set at 300 V and 975 μF. Cells were transferred into complete medium. After 48 hours, 1.0 mg/mL G418 was added to the culture. Individual green fluorescent clones were isolated by limiting dilution into 96-well plates. Specific gene expression was confirmed using reverse transcriptase (RT)-PCR, and protein expression was confirmed by Western blotting using an anti-GFP monoclonal antibody (Covance, Princeton, NJ). Detection of the secreted form of PAI-2 was determined by Western blotting as described under “Immunoprecipitations and Western blotting.”
RT-PCR analysis
RNA from cell cultures was prepared using the Trizol (Invitrogen) method as described previously.20 The purity of the RNA was determined colorimetrically. The SuperScript One-step RT-PCR kit (Invitrogen), with platinum Taq, was used to amplify RNA in the samples. For PAI-2, the primer set used was human PAI-2 forward, 5′-CAGATGAAATTGCCGATGTG-3′, and reverse, 5′-CCATGTCCAGTTCTCCCTGT-3′, which gives a 346-bp (base pair) product, and mouse Pai-2 forward, 5′-CACCACAGGGGGATTATTTG, and reverse, 5′-GGATGCGTCCTCAATCTCAT, which gives a 482-bp product.
Gene expression analysis
RNA extracted from individual HEK-293 cell clones was used to prepare cRNA for hybridization to the Affymetrix U133A oligonucleotide arrays (Affymetrix, Santa Clara, CA) as described previously.20,21 The gene-expression profile from the ZNF198/FGFR1-expressing cells was compared with that obtained from HEK-293 clones stably expressing the GFP protein from the pEGFP vector. To assess gene expression differences, the baseline-corrected data were imported into the Affymetrix Data Mining Tool (DMT 4.0) using the publishing tool, MicroDataBase (MDB 3.0). After the genes were sorted using a count and percentage tool, a list of only those that showed altered expression in at least 2 of the ZNF198/FGFR1 clones (2 cross-comparisons) was compiled. A cutoff of an average 2-fold or greater change was selected. All functional annotation and chromosomal locations were obtained using NetAffx.22
Immunoprecipitations and Western blotting
Cells grown to 80% confluency were washed twice with PBS and lysed in RIPA buffer (50 mM Tris containing 150 mM NaCl, 0.1% SDS, 1% TritonX-100, 1% sodium deoxycholate, pH 7.2) with 0.2% protease and phosphatase inhibitor cocktails (Sigma, St Louis, MO) on ice for 10 minutes. Following centrifugation at 18 000g for 20 minutes the supernatant was collected and used for the analysis of the expression of PAI-2 using Western blotting. When immunoprecipitations (IPs) were performed, the lysates were first passed through rabbit IgG-coupled Sepharose to remove proteins that bind IgG nonspecifically, and then 500 μL supernatant was combined with 3 μL anti-GFP rabbit polyclonal serum and incubated overnight at 4°C with slow mixing (Clontech, Palo Alto, CA). The immune complexes were applied to Protein A-Sepharose and centrifuged. The Sepharose beads were then washed 5 times with PBS, and the proteins were eluted by incubation in SDS-sample buffer at 100°C for 3 minutes. Aliquots were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. For the analysis of secreted PAI-2, 80% confluent cell cultures were washed twice with PBS and incubated overnight in serum-free medium. Supernatants were then collected and concentrated using Amicon (Bedford, MA) ultra centrifugal filters with a molecular weight cutoff of 10 kDa. The concentrated supernatants were analyzed for the presence of PAI-2 by Western blot analysis. Antibodies for PAI-2 were purchased from American Diagnostica, Greenwich, CT.
DNA fragmentation analysis
Cells were plated in 6-well plates and after 24 hours were treated with TNF-α (10 ng/mL) and cycloheximide (1 μg/mL) overnight at 37°C. The cells were then harvested and resuspended in lysis buffer (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 4 mM EDTA, 0.5% SDS, and 0.5% TritonX-100). The cell lysate was incubated with RNase (0.2 mg/mL) overnight at room temperature and then with proteinase K (1 mg/mL) for 1 hour at 37°C. DNA was precipitated with 75% cold ethanol, and the pellet was recovered at 14 000g for 10 minutes and then air-dried and dissolved in 20 μL Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, containing 1 mM EDTA). This preparation was used for DNA fragmentation analysis by electrophoresis through a 1% agarose gel, and the DNA fragments were visualized with ethidium bromide under UV radiation.
Stepwise separation of nuclear and cytoplasmic extracts from cells
Preparation of cytoplasmic and nuclear extracts was achieved by using NE-PER nuclear and cytoplasmic extraction reagents purchased from Pierce, Rockford, IL. Briefly, cells were harvested and washed twice with cold PBS. Cold cytoplasmic extraction reagent I (CER I) was mixed with 0.2% protease inhibitor cocktail (Calbiochem, La Jolla, CA) and added to the cell pellet at a volume of 1 mL CER I/100 μL packed cell volume. The tube was vigorously stirred for 15 seconds, and the cell suspension was left on ice for 10 minutes. Cold cytoplasmic extraction reagent II (CER II; 55μL) was added to the suspension, stirred vigorously for 5 seconds, and left on ice for 1 minute. The tube was again stirred and centrifuged at 16 000g for 5 minutes. The supernatant was collected and used as the cytoplasmic fraction. The insoluble pellet was resuspended in 500 μL cold nuclear extraction reagent (NER) containing 0.2% protease inhibitor cocktail. The tube was stirred for 15 seconds. Sample was left on ice and stirred for 15 seconds every 10 minutes for a total of 40 minutes. The sample was centrifuged at 16 000g, and the supernatant was collected and used as the nuclear extract.
Chromogenic assay for the effect of PAI-2 on uPA activity
The PAI-2 chromogenic assay measures the release of a specific chromophore from its substrate in the presence of uPA. The assay was performed in 96-well plates to determine the activity of uPA in the presence of various culture supernatants. Standard uPA, plus substrate, was used to establish the baseline level of its activity. When commercial, purified PAI-2 is added to this reaction, greater than 99% inhibition is seen. The uPA activity in the presence of various cell-culture supernatants was compared with these standards and expressed as relative OD to assess the level of inhibition. When cell cultures reached 80% confluence, fresh serum-free medium was added, and then these culture supernatants were collected after 24 hours and concentrated using Amicon concentrators. Protein levels were measured using Bradford reagent (Pierce). Different concentrations of the protein were incubated at 37°C in 10 mM HEPES buffer, pH 7.4, containing 5 IU human high molecular weight uPA, 50 μg/mL human Glu-plasminogen, and 1 mM chromogenic substrate, Spectrozyme PL (H-D-norleucyl-hexahydrotyrosol-lysine-para-nitroanilide diacetate). uPA, plasminogen, and Spectrozyme PL were obtained from American Diagnostica. Reactions were performed for 30 minutes, and the release of free p-nitroaniline from the chromogenic substrate was measured at 415 nM using a Bio-Rad Benchmark Plate reader.
Results
The ZNF198/FGFR1 fusion kinase has been shown to regulate the activation of members of the STAT family of transcription factors14 and so clearly affects the gene-expression profile of cells expressing it. We have previously generated HEK-293 cells which stably express a GFP-tagged ZNF198/FGFR1 chimeric gene as well as a wild-type ZNF198 gene.14,18 To investigate the nature of gene expression changes as a result of expression of the ZNF198/FGFR1 fusion protein, we used the Affymetrix U133A oligonucleotide arrays to compare expression profiles between clones from these 2 stable transfectants with those expressing GFP alone. To confirm that the individual clones expressed the correctly sized proteins, we used Western blotting.14,18
Analysis of oligonucleotide microarrays
cRNA from 2 different clones was hybridized to the U133A chips. After baseline correction, the data were arbitrarily filtered to display only those genes that were at least 2-fold up-regulated or down-regulated compared with HEK-293 cells carrying the empty pEGFP vector. This analysis revealed up-regulation of 203 genes and down-regulation of 303 genes in ZNF198/FGFR1-expressing cells. Among the genes showing altered expression levels in ZNF198/FGFR1-expressing cells, many belonged to the serine proteinase inhibitor (SERPIN) gene family (Table 1). For example, pigment epithelium-derived factor (PEDF/SERPINF1), which is known to enhance cellular differentiation, was down-regulated 11-fold in ZNF198/FGFR1-expressing cells. HSP47, (SERPINH1, also called collagen binding protein), was decreased 7-fold in ZNF198/FGFR1-expressing cells compared with control cells. PAI-2, which is not expressed at detectable levels in HEK-293 cells expressing GFP alone, was apparently induced at high levels in ZNF198/FGFR1-expressing cells. PAI-2 is one of the members of the well-known urokinase plasminogen (uPA) activation system, which has been shown to play an important role in tumor growth, invasion, and metastasis. Interestingly, the expression profile of the other components of the uPA system, namely PAI-1 (SERPINE1), uPA, and its receptor (uPAR), was not altered in ZNF198/FGFR1-expressing cells.
Analysis of PAI-2 expression in ZNF198/FGFR1-expressing cells
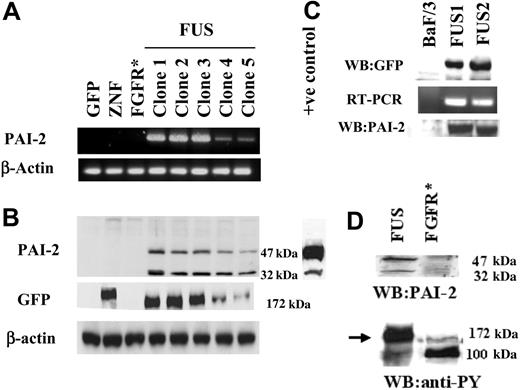
Among the SERPIN family, PAI-2 showed the highest up-regulation in our analysis and has also been shown to be up-regulated in certain myeloproliferative diseases and leukemias.23 This finding prompted us to focus further on the role of PAI-2 up-regulation in ZNF198/FGFR1 fusion kinase-expressing cells. RT-PCR and Western blot analyses were used to analyze HEK-293 cells which expressed GFP, ZNF198, or ZNF198/FGFR1. Detectable levels of PAI-2 were only present in the ZNF198/FGFR1-expressing cells (Figure 1A). Western blot analysis of cell extracts confirmed that only the cells expressing ZNF198/FGFR1 contained the PAI-2 protein (Figure 1B). All of the different clones expressing ZNF198/FGFR1 also expressed PAI-2 in relatively proportional amounts. In our earlier report,14 we observed that ZNF198/FGFR1-expressing cells tended to lose the fusion gene after various lengths of time in culture. This meant that we must constantly subclone to maintain high levels of expression in the overall population. It is likely, therefore, that the 2 clones (4 and 5), which show lower levels of both ZNF198/FGFR1 and PAI-2 expression, are in the process of losing the fusion kinase. Interestingly, all these clones expressed 2 molecular forms of PAI-2; one that was 47 kDa that is expressed in many different cell types, and the other which was approximately 32 kDa. It has been reported previously that PAI-2 is degraded under conditions of induced apoptosis, although the breakdown product still remains active as a uPA inhibitor.24,25 In our experiments, it was apparent, however, that in clones expressing low levels of ZNF198/FGFR1, the reduction of the 32-kDa product was disproportionate relative to the 47-kDa protein.
Analysis of PAI-2 expression. (A) RT-PCR analysis of PAI-2 expression in HEK-293 cells. Clones expressing only GFP, or GFP-tagged exogenous ZNF198 and FGFR1 (* indicates treated with bFGF) do not show detectable levels of PAI-2. In contrast, all 5 independently derived clones expressing ZNF198/FGFR1 (FUS) show PAI-2 expression. Clones 4 and 5 show reduced levels (see text). When these same cell clones were analyzed by Western blotting (B) the absence of PAI-2 was again seen in the cells carrying GFP, GFP-tagged ZNF, and GFP-tagged FGFR1 (bFGF stimulated), but all 5 clones carrying the ZNF198/FGFR1 gene showed 2 size variants of PAI-2, which were 47 and 32 kDa. A positive control from lipopolysaccharide-activated human monocytes was used to confirm the expression of the 2 molecular forms of PAI-2 in ZNF198/FGFR1-expressing clones. This control also demonstrates that the PAI-2 levels in cells expressing the fusion gene are not superphysiologic. When these same samples were probed with an anti-GFP antibody for Western blot analysis, the presence of the 177-kDa ZNF198 protein is detected in cells transfected with GFP-tagged ZNF198 and the smaller 172-kDa ZNF198/FGFR1 protein is seen in all of the 5 clones transfected with GFP-tagged ZNF198/FGFR1. The low levels of ZNF198/FGFR1 protein in clones 4 and 5 parallel the low levels of PAI-2 expression. Samples expressing the GFP vector alone showed only the 27-kDa band (not shown in the figure). (C) BaF/3 clones expressing the ZNF198/FGFR1 gene, detected by Western blotting using anti-GFP, also show induction of the PAI-2 mRNA and protein compared with parental BaF/3 cells, which do not. (D) Addition of bFGF to HEK-293 cells expressing the GFP-tagged FGFR1 gene does not result in the induction of PAI-2, although tyrosine phosphorylation of both the ZNF198/FGFR1 protein (arrow) and FGFR1 in FGFR1-expressing HEK-293 cells (100 kDa) is clearly seen.
Analysis of PAI-2 expression. (A) RT-PCR analysis of PAI-2 expression in HEK-293 cells. Clones expressing only GFP, or GFP-tagged exogenous ZNF198 and FGFR1 (* indicates treated with bFGF) do not show detectable levels of PAI-2. In contrast, all 5 independently derived clones expressing ZNF198/FGFR1 (FUS) show PAI-2 expression. Clones 4 and 5 show reduced levels (see text). When these same cell clones were analyzed by Western blotting (B) the absence of PAI-2 was again seen in the cells carrying GFP, GFP-tagged ZNF, and GFP-tagged FGFR1 (bFGF stimulated), but all 5 clones carrying the ZNF198/FGFR1 gene showed 2 size variants of PAI-2, which were 47 and 32 kDa. A positive control from lipopolysaccharide-activated human monocytes was used to confirm the expression of the 2 molecular forms of PAI-2 in ZNF198/FGFR1-expressing clones. This control also demonstrates that the PAI-2 levels in cells expressing the fusion gene are not superphysiologic. When these same samples were probed with an anti-GFP antibody for Western blot analysis, the presence of the 177-kDa ZNF198 protein is detected in cells transfected with GFP-tagged ZNF198 and the smaller 172-kDa ZNF198/FGFR1 protein is seen in all of the 5 clones transfected with GFP-tagged ZNF198/FGFR1. The low levels of ZNF198/FGFR1 protein in clones 4 and 5 parallel the low levels of PAI-2 expression. Samples expressing the GFP vector alone showed only the 27-kDa band (not shown in the figure). (C) BaF/3 clones expressing the ZNF198/FGFR1 gene, detected by Western blotting using anti-GFP, also show induction of the PAI-2 mRNA and protein compared with parental BaF/3 cells, which do not. (D) Addition of bFGF to HEK-293 cells expressing the GFP-tagged FGFR1 gene does not result in the induction of PAI-2, although tyrosine phosphorylation of both the ZNF198/FGFR1 protein (arrow) and FGFR1 in FGFR1-expressing HEK-293 cells (100 kDa) is clearly seen.
To determine whether expression of ZNF198/FGFR1 induces expression of PAI-2 in hematopoietic cells, as well as HEK-293 cells, we generated BaF/3 clones constitutively expressing the fusion kinase. Consistent with previous reports, in our analysis expression of the fusion kinase results in IL-3 independence in these BaF/3 clones. RT-PCR and Western blot analysis demonstrated that PAI-2 is not expressed endogenously in BaF/3 cells (Figure 1C) but is induced in cells expressing ZNF198/FGFR1. Interestingly, BaF/3 cells expressing ZNF198/FGFR1 showed only the higher molecular form (∼ 47 kDa) of PAI-2 and not the smaller 32-kDa form.
It has been shown that the ZNF198/FGFR1 encodes a protein which has a kinase function similar to that of FGFR1. However, unlike the product of the intact endogenous FGFR1, this chimeric protein is an intracellular (nonmembrane bound), constitutively active kinase which can constitutively activate the STAT signaling pathway.14 To determine whether FGFR1-mediated signaling is responsible for the induction of PAI-2 expression, HEK-293 cells were transfected with a full-length FGFR1 gene, and stable exogenous FGFR1-expressing clones were isolated. When these cells were stimulated with bFGF (10 ng/mL in the presence of 100 U heparin), RNA and protein analysis demonstrated that PAI-2 expression is not induced (Figure 1D). Phosphorylation of FGFR1, however, was observed in these cells following bFGF treatment (Figure 1D), indicating that the FGFR1 protein undergoes standard activation. These results demonstrate that neither ZNF198 expression, nor membrane-initiated FGFR1 activation, alone could induce the expression of PAI-2 in HEK-293 cells. Thus, PAI-2 induction seems to represent a unique consequence of the signaling events associated with the expression of the constitutively active ZNF198-FGFR1 fusion kinase.
Subcellular localization of PAI-2 in ZNF198/FGFR1-expressing cells
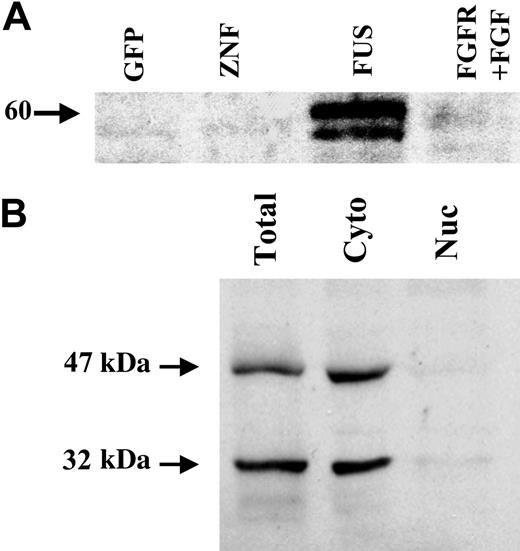
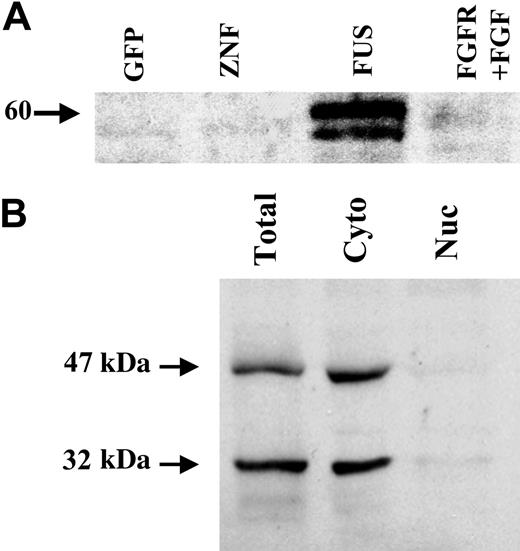
To determine the localization of PAI-2 in cells expressing the ZNF198/FGFR1 fusion gene, the culture supernatants from these cells were analyzed for the presence of the PAI-2 protein. Western blot analysis demonstrated that a 60-kDa glycosylated form of PAI-2 is secreted by ZNF198/FGFR1-expressing cells (Figure 2A) but not by cells expressing the wild-type ZNF198 or GFP alone. PAI-2 was also not secreted in cells expressing FGFR1 after treatment with bFGF. Although it is well established that PAI-2 can be either cytoplasmic or secreted into the extracellular milieu, studies also point to a function for PAI-2 in the nucleus.26 To determine whether this holds true for HEK-293 cells, cytoplasmic and nuclear extracts were prepared using NE-PER extraction reagents (Pierce) and analyzed by Western blotting. These results demonstrated that both the 47-kDa and 32-kDa forms of the PAI-2 protein are predominantly in the cytoplasm in ZNF198/FGFR1-expressing HEK-293 cells (Figure 2B).
Measurement of uPA inhibitory activity in culture supernatants
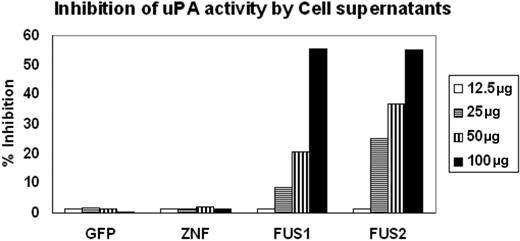
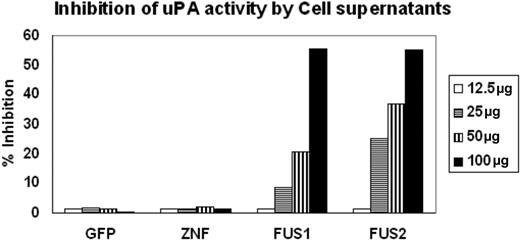
PAI-2 was originally shown to be an inhibitor of urokinase, which converts plasminogen to plasmin. We showed above that PAI-2 is secreted into the culture medium by ZNF198/FGFR1-expressing HEK-293 cells. To further determine whether the PAI-2 protein that is produced in cells expressing the fusion kinase was functional in terms of its activity as a uPA inhibitor, we collected supernatants from HEK-293 cells expressing the ZNF198/FGFR1 chimeric protein and compared their ability to inhibit plasminogen conversion with the similarly processed supernatants from cells expressing either GFP or ZNF198 which do not express PAI-2. The supernatants were concentrated using Amicon concentrators, and chromogenic assay was performed using different concentrations of protein from the supernatants (Figure 3). A fixed concentration of both uPA (5 IU) and plasminogen (50 μg/mL), as well as a fixed 1 mM substrate, was used in each assay. As shown in Figure 3, when increasing concentrations of the culture supernatant was used, no change in chromophore release was seen in the presence of protein obtained from supernatants from GFP- or ZNF198-expressing cells compared with the uPA standard alone. In contrast, there is a dose-dependent increase in the inhibition of uPA-mediated chromophore release in the presence of supernatants from cells expressing ZNF198/FGFR1. These analyses demonstrate that the secreted PAI-2 protein in the cells expressing the fusion kinase can inhibit the function of uPA.
Localization of PAI-2 in HEK-293 cells expressing ZNF198/FGFR1. (A) Analysis of the secretion of PAI-2 into the supernatants from cells transfected with GFP, GFP-tagged ZNF198 (ZNF), GFP-tagged ZNF198/FGFR1 (FUS), or GFP-tagged FGFR1 (after stimulation with bFGF) demonstrates that only those cells expressing ZNF198/FGFR1 secrete PAI-2. (B) Cell lysates from cells expressing the fusion kinase show that the cytoplasmic fraction contains the 47-kDa and 32-kDa forms of PAI-2 but not the nuclear fraction.
Localization of PAI-2 in HEK-293 cells expressing ZNF198/FGFR1. (A) Analysis of the secretion of PAI-2 into the supernatants from cells transfected with GFP, GFP-tagged ZNF198 (ZNF), GFP-tagged ZNF198/FGFR1 (FUS), or GFP-tagged FGFR1 (after stimulation with bFGF) demonstrates that only those cells expressing ZNF198/FGFR1 secrete PAI-2. (B) Cell lysates from cells expressing the fusion kinase show that the cytoplasmic fraction contains the 47-kDa and 32-kDa forms of PAI-2 but not the nuclear fraction.
Coimmunoprecipitation analysis
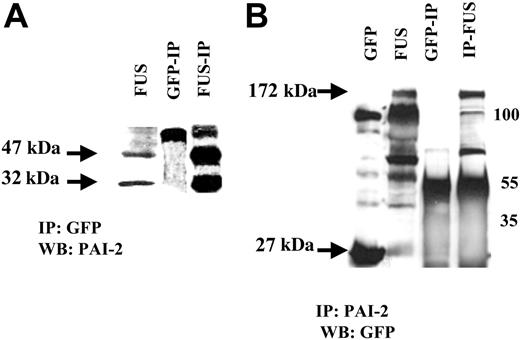
As described above, PAI-2 was induced only in cells that expressed the ZNF198/FGFR1 chimeric protein. This raises the interesting question whether coexpression of the PAI-2 and ZNF198/FGFR1 proteins result in a cooperative interaction. To answer this question, we immunoprecipitated proteins from HEK-293 cells with either an anti-GFP or an anti-PAI-2 antibody and performed Western blot analysis. The results from this analysis demonstrated that PAI-2 binds to the ZNF198/FGFR1 protein (Figure 4) and suggests coincident expression of a proteinase inhibitor, PAI-2, can possibly protect the ZNF198/FGFR1 protein from proteolysis in the cytoplasm.
Analysis of TNF-α-induced cytotoxicity in ZNF198/FGFR1 HEK-293 cells
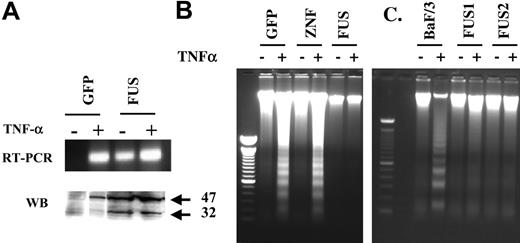
TNF-α has been shown to induce the expression of PAI-2 on its own27,28 and also has diverse effects on cells, some of which are related to PAI-2 expression. For example, in the absence of new protein synthesis following treatment of cells with cycloheximide, PAI-2 induces resistance to TNF-α-mediated cytotoxicity.29 We therefore determined whether PAI-2 expression in ZNF198/FGFR1-expressing cells correlates with resistance of these cells to TNF-α-mediated cytotoxicity. Treatment of GFP-expressing HEK-293 cells with TNF-α alone induced the expression of PAI-2 (Figure 5A). These cells, however, contained only the 47-kDa form of the protein in the cytoplasm. In contrast, the ZNF198/FGFR1-expressing cells possessed both the 47-kDa as well as a 32-kDa proteins in the cytoplasm. TNF-α treatment, in the presence of cycloheximide (1 μM), induced a potent apoptosis in both HEK-293 cells expressing either GFP or ZNF198 (Figure 5B) but not in ZNF198/FGFR1-expressing cells. BaF/3 cells expressing ZNF198/FGFR1 also showed a resistance to TNF-α-mediated apoptosis that was not present in the parental BaF/3 cells (Figure 5C). This analysis demonstrates that PAI-2 expression in ZNF198/FGFR1-expressing cells correlates with resistance to TNF-α-mediated apoptosis.
Inhibition of UPA activity by PAI-2. Inhibition of uPA activity was assessed using the chromogenic assays as described in “Methods and materials.” Supernatants from cells showing forced expression of exogenous GFP or ZNF198 showed virtually undetectable inhibition of uPA activity. In contrast, supernatants from 2 different cell clones expressing the ZNF198/FGFR1 fusion kinase (FUS) show inhibition of uPA activity proportional to the levels of the input protein.
Inhibition of UPA activity by PAI-2. Inhibition of uPA activity was assessed using the chromogenic assays as described in “Methods and materials.” Supernatants from cells showing forced expression of exogenous GFP or ZNF198 showed virtually undetectable inhibition of uPA activity. In contrast, supernatants from 2 different cell clones expressing the ZNF198/FGFR1 fusion kinase (FUS) show inhibition of uPA activity proportional to the levels of the input protein.
Interaction between PAI-2 and ZNF198/FGFR1. HEK-293 cells expressing ZNF198/FGFR1 (FUS) also express the 47-kDa and 32-kDa forms of PAI-2. Coimmunoprecipitation from ZNF198/FGFR1-expressing cells using the GFP antibody (A) identifies the 47-kDa and 32-kDa PAI-2 proteins which are not present in HEK-293 cells expressing GFP alone. The larger 55-kDa band in the IPs is derived from the IgG. (B) The HEK-293 cells expressing GFP (27 kDa) show a nonspecific band at 100 kDa but not the 172-kDa specific band corresponding to the ZNF198/FGFR1 protein (FUS). ZNF198/FGFR1-expressing HEK-293 cells show the fusion protein (172 kDa) and a series of degradation products we described previously.14 Co-IP with an anti-PAI-2 antibody reveals the 172-kDa ZNF198/FGFR1 protein in cells expressing this fusion kinase gene. Smaller bands in these IPs represent IgG heavy chain band and degradation products.
Interaction between PAI-2 and ZNF198/FGFR1. HEK-293 cells expressing ZNF198/FGFR1 (FUS) also express the 47-kDa and 32-kDa forms of PAI-2. Coimmunoprecipitation from ZNF198/FGFR1-expressing cells using the GFP antibody (A) identifies the 47-kDa and 32-kDa PAI-2 proteins which are not present in HEK-293 cells expressing GFP alone. The larger 55-kDa band in the IPs is derived from the IgG. (B) The HEK-293 cells expressing GFP (27 kDa) show a nonspecific band at 100 kDa but not the 172-kDa specific band corresponding to the ZNF198/FGFR1 protein (FUS). ZNF198/FGFR1-expressing HEK-293 cells show the fusion protein (172 kDa) and a series of degradation products we described previously.14 Co-IP with an anti-PAI-2 antibody reveals the 172-kDa ZNF198/FGFR1 protein in cells expressing this fusion kinase gene. Smaller bands in these IPs represent IgG heavy chain band and degradation products.
Discussion
The ZNF198/FGFR1 fusion gene,9 that is associated with an atypical MPD, generates a chimeric protein with constitutive intracellular tyrosine kinase activity. Because the t(8;13) translocation that produces this fusion kinase gene occurs often as the only cytogenetic change in the leukemic cells, it is assumed that the fusion gene acts as a dominant oncogenic event. As a consequence of the expression of the ZNF198/FGFR1 chimeric protein, activity of various STAT proteins is up-regulated,14,19 which presumably restructures the gene-expression profile in the transformed cells. Unfortunately, no cell lines have yet been derived from the MPD which carries the t(8;13) rearrangement. As we reported previously,14 many cell types do not easily tolerate the expression of this powerful oncogenic kinase, although other groups as well as ours have used cell systems such as Cos7, HepG2, BaF3, MCF-7, HEK-293 to study the signal transduction mediated by the ZNF198/FGFR1 fusion kinase.14,19 It was also demonstrated that various STAT gene family members are constitutively activated in these cell types in the presence of the fusion kinase, demonstrating that the function of ZNF198/FGFR1 is not cell type specific. Because HEK-293 cells do not express PAI-2 under normal conditions, we used these cells to investigate the induction of PAI-2 by ZNF198/FGFR1 fusion kinase. As a result of gene-expression profiling we demonstrated that PAI-2, a proteinase inhibitor that has been shown to have multiple functions in preventing targeted proteolytic degradation, was induced in the presence of ZNF198/FGFR1. The same was true for BaF/3 cells. In most cells a balance between proteases and their inhibitors is essential for the orderly turnover of proteins within the cell, which in turn determines the functional state of the cell. An association between changes in the intracellular balance of various proteases and their inhibitors is well documented in diseases such as cancer.30
Analysis of apoptosis induced by TNF-α. Treatment of HEK-293 cells carrying the GFP gene alone (A) with TNF-α in the presence of cycloheximide demonstrates induction of transcription of the PAI-2 gene but only the presence of the 47-kDa protein product. Treatment of cells expressing ZNF198/FGFR1 (FUS), which express both cytoplasmic forms of PAI-2 endogenously, with TNF-α has only a marginal effect on RNA and protein levels of PAI-2. When nucleosome laddering, an indicator of apoptosis, is analyzed in the same cells (B) it can be seen that TNF-α induces apoptosis in cells expressing GFP or the exogenous ZNF198 gene (ZNF), but not in cells expressing the ZNF198/FGFR1 gene (FUS). Similarly, BaF/3 cells (C) expressing the fusion kinase are resistant to TNF-α-induced apoptosis compared with the parental cells which are susceptible.
Analysis of apoptosis induced by TNF-α. Treatment of HEK-293 cells carrying the GFP gene alone (A) with TNF-α in the presence of cycloheximide demonstrates induction of transcription of the PAI-2 gene but only the presence of the 47-kDa protein product. Treatment of cells expressing ZNF198/FGFR1 (FUS), which express both cytoplasmic forms of PAI-2 endogenously, with TNF-α has only a marginal effect on RNA and protein levels of PAI-2. When nucleosome laddering, an indicator of apoptosis, is analyzed in the same cells (B) it can be seen that TNF-α induces apoptosis in cells expressing GFP or the exogenous ZNF198 gene (ZNF), but not in cells expressing the ZNF198/FGFR1 gene (FUS). Similarly, BaF/3 cells (C) expressing the fusion kinase are resistant to TNF-α-induced apoptosis compared with the parental cells which are susceptible.
PAI-2 belongs to the subgroup of serpins known as the ovalbumin-like serine proteinase inhibitors.31-33 Within this subgroup, it is a member of the uPA inhibitor system that converts plasminogen to plasmin. This event often facilitates cell migration and invasion by both degrading fibrin and activating metalloproteinases.34,35 The uPA system also plays a major role in tumor growth, invasion, and metastasis.36,37 In leukemias, it has been shown that PAI-2 is up-regulated in M2 and M4 to M5 types and is thought to be responsible for the invasive phenotype of these cells.23 Consistent with these observations we have also shown that hematopoietic cells forced to express ZNF198/FGFR1 also up-regulate PAI-2. In a report, PAI-2 was also shown to bind to the retinoblastoma (RB1) protein and protect it from degradation by an as yet unknown protease.26 However, because PAI-2 was only detectable in the cytoplasm in the HEK-293 cells that expressed ZNF198/FGFR1, it is possible that its normal nuclear function is disrupted in these cells or it is also possible that the RB1-stabilizing function is performed by a protein other than PAI-2 in HEK-293 cells. Nevertheless, because PAI-2 also binds to the ZNF198/FGFR1 fusion protein in the cytoplasm, it is possible that, as for RB1, PAI-2 protects the fusion kinase from proteolysis. Thus, in MPD cells, preventing the degradation of ZNF198/FGFR1 may be an important mechanism for maintaining the malignant phenotype. PAI-2 is expressed in many normal cell types such as activated monocytes, macrophages, differentiating keratinocytes, and placental trophoblasts and can be regulated by various growth factors, cytokines, hormones, and tumor promoters.34 The PAI-2 protein has so far been reported to exist in 2 forms; an intracellular 47-kDa protein and an extracellular secreted form, which as a result of glycosylation, is 60 kDa. Both of these forms are expressed in ZNF198/FGFR1 kinase-expressing HEK-293 cells but neither is expressed in normal HEK-293 cells. It was reported earlier that PAI-2 is cleaved to a smaller sized product of approximately 33 kDa under conditions of induced apoptosis.24,25 Physiologic relevance of this breakdown product of PAI-2 is not known. In our studies, we have now identified that ZNF198/FGFR1-induced PAI-2 is also cleaved into a 32-kDa form in HEK-293 cells expressing ZNF198/FGFR1 but is not cleaved in BaF/3 cells expressing the fusion kinase. It was also observed that the 32-kDa breakdown product coexists with the 47-kDa form in the cytoplasm. Interestingly, it was observed that, in clones expressing low levels of ZNF198/FGFR1, the relative amount of the 32-kDa cleavage product was disproportionate. These results suggest an important correlation between the stability of the fusion protein and the breakdown of PAI-2. Co-IP experiments demonstrated that both the 47-kDa and 32-kDa forms bind to the fusion kinase, suggesting an active role for the 32-kDa form in protecting it from degradation. When GFP-expressing HEK-293 cells were treated with TNF-α, a well-known transcriptional activator of PAI-2, only the more usual 47-kDa form of PAI-2 was induced. In addition to its role in protein stabilization, PAI-2 also confers resistance to TNF-α-mediated apoptosis.29 The ability to resist apoptosis is an important feature of cancer cells and is particularly relevant in leukemia cells because programmed turnover of normal B and T cells at the end of their designated lifespan usually occurs through apoptosis. Overexpression of BCL2 is often the mechanism by which many leukemic cells are thought to avoid apoptosis, but our GeneChip data demonstrated that BCL2 was not up-regulated in cells expressing the ZNF198/FGFR1 fusion gene. We have now demonstrated that HEK-293 cells as well as BaF/3 cells expressing ZNF198/FGFR1 have become resistant to TNF-α-induced apoptosis in the presence of PAI-2.
In our studies, ZNF198/FGFR1 kinase is mostly sequestered to the cytoplasm, unlike ZNF198 which is present exclusively in the nucleus. The chimeric kinase has also lost the ability to interact with several of the binding partners that otherwise bind to ZNF198.38 Current data that PAI-2 interacts with the ZNF198/FGFR1 kinase, and presumably stabilizes it in the cell, indicate that a stable expression of certain oncogenes may require proteins such as PAI-2 for specific and sustained induction of disease.
Prepublished online as Blood First Edition Paper, January 12, 2006; DOI 10.1182/blood-2005-04-1505.
Supported by the National Institute of Health (grant CA 76167) and in part by the Roswell Park Cancer Institute Cancer Center (support grant CA 16056).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr D. Ginsburg, Howard Hughes Medical Institute, University of Michigan Medical School, for kindly providing the antibody against the mouse PAI-2 protein. We thank Dr Heinz Baumann for reagents supporting the activated monocyte experiments.