Abstract
Severe combined immunodeficiency (SCID) caused by mutations in RAG1 or RAG2 genes is characterized by a complete block in T- and B-cell development. The only curative treatment is allogeneic hematopoietic stem cell transplantation, which gives a high survival rate (90%) when an HLA-genoidentical donor exists but unsatisfactory results when only partially compatible donors are available. We have thus been interested in the development of a potential alternative treatment by using retroviral gene transfer of a normal copy of RAG1 cDNA. We show here that this approach applied to RAG-1-deficient mice restores normal B- and T-cell function even in the presence of a reduced number of mature B cells. The reconstitution is stable over time, attesting to a selective advantage of transduced progenitors. Notably, a high transgene copy number was detected in all lymphoid organs, and this was associated with a risk of lymphoproliferation as observed in one mouse. Altogether, these results demonstrate that correction of RAG-1 deficiency can be achieved by gene therapy in immunodeficient mice but that human application would require the use of self-inactivated vector to decrease the risk of lymphoproliferative diseases.
Introduction
Severe combined immunodeficiency (SCID) is a heterogeneous group of rare autosomic recessive disorders occurring in 1 in 75 000 to 1 in 100 000 births. The common characteristic of these pathologies is the absence of T cells associated with an impaired function of B lymphocytes leading to death within the first year of life in the absence of treatment.1,2 Currently, the treatment of choice is allogeneic hematopoietic stem cell transplantation (HSCT) with a survival rate approximating 90% when an HLA-genoidentical donor is available.3 In most cases, haploidentical donors or unrelated donors are the only source of allogeneic hematopoietic stem cells, and in this case the survival rate ranges from 60% to 75%. Within the SCID subsets, the T-cell-negative, B-cell-negative, natural killer cell-positive (T-B-NK+) group has a poorer prognosis, especially when a partially compatible donor is used (with a survival rate close to 36%). Of note, this subgroup of mismatched patients receiving transplants presents a number of long-term complications due to a frequent persistence of B-cell deficiency and a long-term decline in T-cell functions related to the absence of donor stem cell engraftment.4 The slow kinetics of the T-lymphocyte development also accounts for many lethal infections observed in the first 6 months after HSCT. These limitations justify the search for an alternative strategy to improve the prognosis of SCID. Transfer of a normal copy of the mutated gene that causes the SCID disease to early hematopoietic progenitors has largely been considered, and this therapeutic approach has provided efficient long-term correction in 2 human SCID diseases5-8 as well as in various immunodeficient mouse models.9-12
The T-B-NK+ group of SCID disease represents 20% to 30% of the all SCID forms. Within this subset, mutations in 1 of the 2 recombination activating genes, RAG1 or RAG2,13,14 account for half of the cases. Both RAG-1 and RAG-2 proteins play a key role in the initiation of the V(D)J recombination step, crucial for the formation of the antigen receptor at the surface of T and B lymphocytes.15,16 The simultaneous expression of RAG-1 and RAG-2 is required during the early stages of T- and B-lymphocyte maturation and, in the absence of 1 of the 2 proteins, lymphoid differentiation is blocked at the pro-B and triple-negative pre-T stages.17 Of note, in humans, hypomorphic mutations of RAG proteins can lead to a leaky phenotype, the Omenn syndrome, characterized by the presence of a few autoreactive T-cell clones.18 RAG-1- and RAG-2-deficient mice (RAG-1-/- and RAG-2-/-, respectively) exhibit an identical phenotype19,20 to the one observed in RAG-deficient patients with null mutations. Therefore, these animals represent a good model to test the efficiency and toxicity of RAG1 gene therapy approach.
We have previously demonstrated that RAG-2 deficiency could be long-term corrected following retroviral transfer of the human RAG2 gene in the HSCs of RAG-2-/- mice.11 Using the same strategy, the human RAG1 (hRAG1) transgene was cloned in a Moloney leukemia virus (MLV) vector used to transduce bone marrow progenitors of RAG-1-/--deficient mice. T- and B-cell differentiation was restored in all the lymphoid organs for more than 1 year after transplantation provided that the transgene copy number per cell was high enough. Of note, such a high provirus copy number leads to the occurrence of one leukemic event. Together with the observed clinical complications in the γc deficiency gene-therapy trial21 these results indicate that alternative transduction methods, such as long-terminal repeat (LTR)-inactivated retroviral vectors, should be used for this therapeutic approach.
Materials and methods
Mice
Four- to 6-week-old C57BL/10 wild-type mice and C57BL/10 RAG-1-deficient mice were obtained from Charles River Laboratories (L'Arbresle, France). The animals were maintained in a specific pathogen-free animal facility. All experiments and procedures were performed in compliance with the French Ministry of Agriculture Regulations for Animal Experimentations (Act no. 87847, October 19, 1987, modified May 2001).
Retroviral supernatant production
MFG-RAG1 vector was obtained by a SalI/NcoI ligation of the human RAG1 cDNA product into the MFGB2 plasmid, which has been described elsewhere.22 Retroviral supernatant was produced in Stemspan medium (StemCell Technologies, Vancouver, BC, Canada) supplemented with 10% fetal bovine serum (FBS; StemCell Technologies). Briefly, ecotropic phoenix producer cell lines were transfected following a calcium-phosphate protocol as previously described.11 After 48 hours, the supernatant was collected and concentrated onto vivaspin concentrator tubes (Vivascience, Hannover, Germany). Virus titration was performed on 3T3 cell lines, estimated at 6 × 106 infectious particles per milliliter and used at a multiplicity of infection of 3. The detection of replication-competent retrovirus (RCR) in the retroviral supernatant batches was performed by Texcell (Paris, France).23 No RCRs were detected.
Transduction of bone marrow cells
Bone marrow cells were flushed from both tibias and femora of donor mice and treated with a 0.75% NH4Cl (Sigma-Aldrich, St Louis, MO) solution in Tris (tris(hydroxymethyl)aminomethane buffer) (Sigma-Aldrich) to remove red blood cells (RBCs). Cells were stained with phycoerythrin (PE)-conjugated Sca-1 Ly6A/E (E13-161-7) antibody (Becton Dickinson [BD] Biosciences Pharmingen, San Jose, CA), and Sca-1+ cells were purified using an anti-PE magnetic selection kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Sca-1+ cells were cultured for 24 hours in Stemspan medium (StemCell Technologies) supplemented with 5% FBS (StemCell Technologies) and the following recombinant cytokines: murine stem cell factor (mSCF) 100 ng/mL (Abcys, Paris, France), human FMS-like tyrosine kinase 3-ligand (hFlt3-L) 100 ng/mL and human megakaryocyte growth and development factor (hMGDF) 100 ng/mL (Amgen, Thousand Oaks, CA), mIL-6 50 ng/mL (Abcys), and mIL-11 10 ng/mL (R&D, Minneapolis, MN). Cells were seeded at 1 × 106/mL in flat-bottomed P96 wells coated with 50 μg/mL recombinant human fibronectin fragment (RetroNectin CH-296; Takara Biomedicals, Shiga, Japan). Three transduction cycles were performed at 24-hour intervals by replacing the medium with retroviral supernatant supplemented with the same cytokines and protamine sulfate (4 μg/mL) (Choay, Gentilly, France). One million cells were injected intravenously into RAG-1-deficient mice previously irradiated at 300 or 800 cGy using a 137Cs irradiator. The control group was obtained by transplanting 1 × 106 wild-type C57BL/10 cells previously cultured with the protocol described for transduced cells. Mice treated with MFG-RAG-1 were killed 4 to 6 months after transplantation, and secondary grafts were performed as follows: bone marrow cells were collected, treated with an NH4Cl solution, and a total of 5 × 106 cells per milliliter were injected intravenously to 300 cGy- or 800 cGy-irradiated RAG-1-/- recipients.
Flow cytometry analysis
Flow cytometry analyses were performed on cells obtained from blood, thymus, bone marrow, spleen, and lymph nodes. Cells were counted with trypan blue and stained with the following rat anti-mouse monoclonal antibodies (MoAbs) (BD Biosciences Pharmingen): fluorescein isothiocyanate (FITC)-conjugated CD3 (145-2C11), FITC-conjugated CD25 (PC61), PE-conjugated CD8 (53-5.8), PE-conjugated CD45R/B220 (RA3-6B2), allophycocyanin (APC)-conjugated CD4 (L3T4), APC-conjugated T-cell receptor (TCRβ) (H57-597), APC-conjugated CD44 (IM7), a polyclonal goat anti-mouse IgM FITC conjugated (Jackson ImmunoResearch Labs, West Grove, PA), and a rat anti-mouse IgD PE conjugated (11-26; Southern Biotechnology Associates, Birmingham, AL). To prevent possible binding to Fc receptors, peripheral blood cells were preincubated with anti-mouse CD16/CD32 monoclonal antibodies (MoAbs) (2-4G2; BD Biosciences Pharmingen) and treated, after staining, with fluorescence-activated cell sorter (FACS) lysing solution (BD Biosciences Pharmingen) to remove RBCs. Analyzes were performed on a FACSCalibur (BD Biosciences Pharmingen) using Cellquest software.
Lymphocyte proliferation assays
Spleen or lymph node cells were cultured at 1 × 106/mL in supplemented Dulbecco modified Eagle medium (DMEM; GibcoBRL, Rockville, MD) with 10% FBS (GibcoBRL) for 3 days in the presence of anti-CD3 (2-C11; 10 μg/mL) and anti-CD28 (37.51; 1 μg/mL) antibodies (BD Biosciences Pharmingen) or for 4 days in the presence of lipopolysaccharide (LPS; 50μg/mL; Sigma-Aldrich). Proliferation was assayed by incorporation of [3H]thymidine (2 μCi/mL [0.074 MBq]; Amersham, Saclay, France). After overnight incubation, cells were transferred onto filters and placed in 1 mL scintillation liquid (Packard Biosciences, Groningen, The Netherlands). Uptake of [3H]thymidine was determined with a scintillation counter (Packard Biosciences).
Analysis of TCR repertoire
Total RNA was prepared from splenocytes using the Rneasy kit (Qiagen, Valencia, CA), and cDNA was synthesized using the expand reverse transcriptase (Roche, Indianapolis, IN). For TCRVβ quantitative immunoscope analysis, cDNA was amplified with each of 24 TCRVβ family-specific primers together with a TCRCβ primer and an Minor Groove Binder (MGB)-TaqMan probe for TCRCβ. For TCRVα quantitative immunoscope analysis, cDNA was amplified with each of 21 TCRVα family-specific primers together with a TCRCα primer and an MGB-TaqMan probe for TCRCα. Real-time quantitative polymerase chain reaction (PCR) was carried out on the ABI7300 system (Applied Biosystems, Foster City, CA). PCR products were then subjected to runoff reactions by using nested fluorescents primers specific for the Cβ or Cα segment. Labeled products were resolved on an automated 373A sequencer (Perkin Elmer, Foster City, CA). The fluorescence intensity of each band was recorded and analyzed using Immunoscope software (Perkin Elmer).24,25
Serum immunoglobulin quantification and antibody response to a T-cell-dependent antigen
IgM and IgG immunoglobulin levels were obtained by quantitating serial dilutions of serum samples with an enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions (Bethyl Laboratories, Montgomery, TX). T-cell-dependent responses against keyhole limpet hemocyanin (KLH) were tested following immunization of mice intraperitoneally with 50 μg KLH (Sigma-Aldrich) solution in alum (Sigma-Aldrich). Mice were boosted with a 50-μg KLH intraperitoneal injection 21 days after immunization, and serum samples were drawn 28 days after immunization. Anti-KLH-specific immunoglobulins were detected by precoating the wells with KLH (50 μg/mL) before ELISA detection.
Quantification of genome proviral integrations and expression
Integration study was performed on RAG1-transduced Sca-1+ cells and on lymphoid and granulocyte lineage subsets isolated from spleen cells. Briefly, splenocytes from RAG1-transduced mice were marked with PE-conjugated CD4 and CD8 Abs (145-2C11; BD Biosciences Pharmingen) and APC-conjugated GR-1 (Ly6-G; BD Biosciences Pharmingen) or by APC-conjugated B220 (RA3-6B2; BD Biosciences Pharmingen) and polyclonal PE-conjugated IgM (Jackson ImmunoResearch Labs). Gr-1+, CD4+/CD8+, and B220+IgM+ cells were sorted with a FACStar (BD Biosciences Pharmingen) with a purity of at least 99% for each subset. Genomic DNA was extracted by phenol/chloroform extraction. Real-time quantitative PCR was performed with the ABI Prism 7700 Sequence Detector System (Applied Biosystems). Amplification conditions consisted of an AmpliTaq Gold activation step at 95°C for 10 minutes followed by 40 cycles of 2 steps: 15 seconds of denaturation at 95°C and 60 seconds of annealing at 60°C. TaqMan probes were used for detection of the human RAG1 transgene and a sequence specific of the murine genome (a nonrepetitive sequence of the fifth exon of the M region of titin). For each primer set, a standard curve was generated by serial dilution of a DNAplasmid containing the sequence amplified. The data were calculated using the standard curve method giving PCR efficiency for each primer set and attributing values for each sample relative to the reference sample control. All samples and serial dilutions were run in duplicate. The reference control was a plasmid containing both sequences (titin and RAG1), and the results obtained from titin quantification were used to normalize the number of copies back to the number of cells.
The primers and probe sequences used are as follows: 1894MFGRAG1. forward, GGTGGACCATCCTCTAGACTGC; 1979MFGRAG1.reverse, TGGGTGCTGAATTTCATCTGG; 1920MFGRAG1.probe, CAGCCTCTTTCCCACCCACCTTGG; m139Mex5.forward, AAAACGAGCAGTGACGTGAGC; m261Mex5.reverse, TTCAGTCATGCTGCTAGCGC; M161TitinMex5.probe, TGCACGGAAGCGTCTCGTCTCAGTC.
hRAG1 transgene expression was quantified in the samples obtained from various tissues like spleen, thymus, bone marrow, or lymph nodes. RNA was extracted with the Rneasy mini kit (Qiagen, Hilden, Germany). One microgram of total RNA was reverse transcribed to single-stranded cDNA with 50 units of Superscript II Reagent (Invitrogen, Carlsad, CA) and random hexamers. Reverse transcription (RT) was performed in a 20-μL final volume, and the sample was incubated at 42°C for 60 minutes. To account for variations due to RNA extraction and the RT reaction, the measured level of hRAG1 transgene mRNA was normalized against an endogenous RNA control, PO mRNA, an acidic ribosomal phosphoprotein ubiquitously expressed in all murine cells.26 Ratios were calculated by comparing the hRAG-1 mRNA expression levels (hRAG-1/PO) in the tested samples with murine RAG-1 expression levels in the thymus of wild-type C57BL/10 mice (mRAG-1/PO). The hRAG-1 transcript expression value was calculated as a percentage of the reference mRAG-1/PO ratio in the thymus: % hRAG-1 expression = (hRAG-1/PO)/(mRAG-1/PO) × 100.
The primers and probe sequences used are as follows: 678hRAG1. forward, TCAGCCAAACTTGCAGCTCA; 780hRAG1.reverse, CTTGCTGCTGATCCTTGCCT; 722hRAG1.probe, ACCAAGCAAGACAAGCCCGTCAGC; MH181PO.forward, CTCCAAGCAGATGCAGCAGA; M267PO. reverse, ATAGCCTTGCGCATCATGGT; M225PO.probe, CCGTGGTGCTGATGGGCAAGAA.
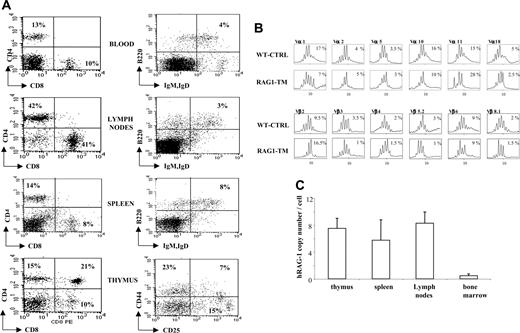
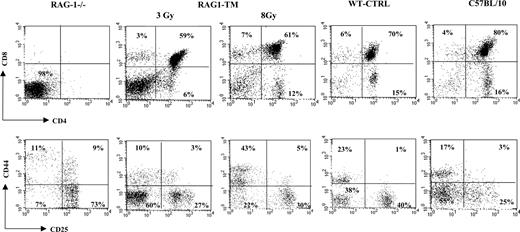
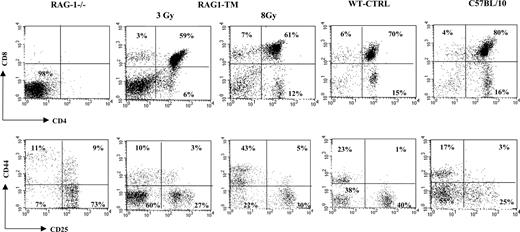
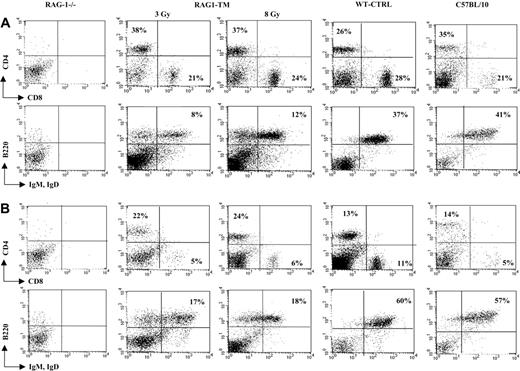
Thymus profile after RAG1 retroviral-mediated gene transfer into RAG-1-deficient mice. Mice receiving transplants were killed 6 months after RAG1 gene transfer. Flow cytometry analysis was performed on total thymic cells for CD4 and CD8 staining. For the CD25 and CD44 distribution analysis, the dot-plot window was gated on the CD4-CD8- cells. The dot plots are representative of one experiment: n = 4 for the RAG-1-deficient mice (RAG-1-/-), n = 5 for the RAG1-transduced mice (RAG1-TM, 3 Gy), n = 4 for the RAG1-transduced mice (RAG1-TM, 8 Gy), n = 8 for the control mice (WT-CTRL), and n = 4 for the same age wild-type C57BL/10 mice. Percentages represent the proportion of cells expressing CD4, CD8, CD25, or CD44 markers.
Thymus profile after RAG1 retroviral-mediated gene transfer into RAG-1-deficient mice. Mice receiving transplants were killed 6 months after RAG1 gene transfer. Flow cytometry analysis was performed on total thymic cells for CD4 and CD8 staining. For the CD25 and CD44 distribution analysis, the dot-plot window was gated on the CD4-CD8- cells. The dot plots are representative of one experiment: n = 4 for the RAG-1-deficient mice (RAG-1-/-), n = 5 for the RAG1-transduced mice (RAG1-TM, 3 Gy), n = 4 for the RAG1-transduced mice (RAG1-TM, 8 Gy), n = 8 for the control mice (WT-CTRL), and n = 4 for the same age wild-type C57BL/10 mice. Percentages represent the proportion of cells expressing CD4, CD8, CD25, or CD44 markers.
LAM-PCR analysis
An integration site restriction fragment length display was obtained by linear amplification-mediated PCR (LAM-PCR) consisting of repeated primer extension, second strand synthesis, restriction digest, cassette ligation, and exponential amplification as described previously.27 LAM-PCR was performed on DNA isolated from spleen, bone marrow, or liver cells, and the final product was separated on Spreadex gels (Elchrom, Cham, Switzerland) to visualize the polymorphism of integrations sites.
Statistical analysis
Mann-Whitney U tests were used to compare immune reconstitution between RAG1-transduced mice and control mice. Data were considered to be statistically significant at P values less than .05.
Results
Immune reconstitution following ex vivo RAG1 gene transfer
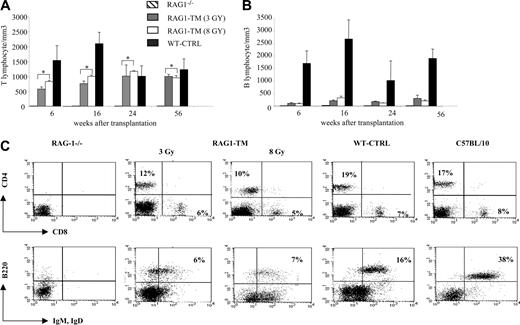
Six months following gene transfer, T-cell development was analyzed in the RAG1-transduced mice (RAG1-TM, n = 9) as well as in RAG-1-/- control mice that received congenic wild-type bone marrow cells (WT-CTRL, n = 8). In the recipient mice irradiated with a 8 Gy dose (n = 4), thymocyte counts and thymic reconstitution feature are undistinguishable from WT-CTRL mice (Figure 1; Table 1). In the 3 Gy-irradiated group (n = 5), overall thymocyte counts were more variable (from 1 × 105 cells to 8 × 106 cells) and lower than the thymocyte counts observed in the 8 Gy-irradiated group (P < 5%). Of note, CD4+CD8+ double-positive (DP) cells were found in only 2 of 5 recipients although mature single-positive (SP) thymocytes (CD4+CD3+ or CD8+CD3+) were detected in all cases. In both RAG1-TM and WT-CTRL groups, the presence of DP cells was associated with a normal distribution of the double-negative thymocyte subsets as studied by the CD25 and CD44 surface antigen expression (Figure 1). Together, these data demonstrate that thymus reconstitution following ex vivo retroviral gene transfer is directly dependent on the recipients' irradiation dose. The spleen, lymph node, and bone marrow cell counts were similar in all groups—that is, RAG1-TM (3 or 8 Gy irradiated), WT-CTRL, and wild-type C57BL/10 mice (Table 1). However, the proportion of mature B lymphocytes (B220+IgM+IgD+) was reduced in the lymphoid organs of RAG1-TM mice when compared with both control groups. Such a difference was not observed in the mature T-cell compartment (Figure 2). In all the RAG1-TM mice, circulating mature T cells became detectable 6 weeks after gene therapy with a slight steady increase during the first 4 months of reconstitution. T-cell counts and the CD4/CD8 ratio reached WT-CTRL levels within 6 months and remained stable up to 1 year after gene therapy (Figure 3A,C). These cell counts were very similar to those detected in age-matched wild-type C57BL/10 mice, attesting that the T-cell peripheral pool was fully reconstituted. In contrast, blood B-lymphocyte reconstitution was delayed in RAG1-TM mice, and B-cell counts remained persistently lower than in control mice (Figure 3B-C). Of note, B-cell counts in WT-CTRL mice were lower than in age-matched wild-type C57BL/10 mice (1883 ± 372 cells/mm3 and 4300 ± 809 cells/mm3, respectively) even 1 year after gene therapy. Unlike the differences observed between thymic reconstitution in the 3 Gy- and 8 Gy-recipient mice, cell counts and lymphocyte subset distribution were similar in the lymphoid organs and in the peripheral blood of both irradiated recipient groups.
T- and B-lymphocyte reconstitution. Reconstitution is depicted in lymph nodes (A) and spleen (B) following RAG1 gene therapy. Six months after transplantation, mice were killed, and flow cytometry analysis was performed to measure the proportion of mature T and B lymphocytes in the different organs. The dot plots are representative of 1 of 9 RAG1-TM (n = 5 for 3 Gy and n = 4 for 8 Gy), 8 WT-CTRL, 4 C57BL/10, and 4 RAG-1-/- mice that gave similar results. Percentages represent the proportion of cells expressing CD4, CD8, B220, IgM, or IgD markers.
T- and B-lymphocyte reconstitution. Reconstitution is depicted in lymph nodes (A) and spleen (B) following RAG1 gene therapy. Six months after transplantation, mice were killed, and flow cytometry analysis was performed to measure the proportion of mature T and B lymphocytes in the different organs. The dot plots are representative of 1 of 9 RAG1-TM (n = 5 for 3 Gy and n = 4 for 8 Gy), 8 WT-CTRL, 4 C57BL/10, and 4 RAG-1-/- mice that gave similar results. Percentages represent the proportion of cells expressing CD4, CD8, B220, IgM, or IgD markers.
T- and B-cell characteristics after gene therapy
To analyze the Vβ and Vα diversity of the TCR, immunoscope studies were performed on splenocytes 6 months after transplantation. No significant differences were observed between RAG1-TM and WT-CTRL mice (Figure 4A-B), indicating that RAG1 gene transfer restored a T-lymphocyte pool with a diverse T-cell repertoire. To assess the functionality of the emerging T lymphocytes, proliferation assays were performed on lymph node-derived cells stimulated with anti-CD3+anti-CD28 moAbs. As shown in Figure 4C, a similar proliferative response was detected in T cells from both RAG1-TM and WT-CTRL animals. We then assessed what consequences the low level of B-lymphocyte reconstitution had on B-cell function. Proliferation assays were performed by stimulating splenocytes with LPS for 4 days. Similar B-cell proliferations were detected in both RAG1-TM and WT-CTRL animals (Figure 4D). Total IgG and IgM serum immunoglobulin levels were found to be equivalent in RAG1-TM and WT-CTRL mice (Figure 4E). Moreover, following immunization with the T-dependent antigen KLH, newly generated B cells were able to produce specific antibodies of both IgM and IgG isotypes to a similar extent as WT-CTRL (Figure 4F).
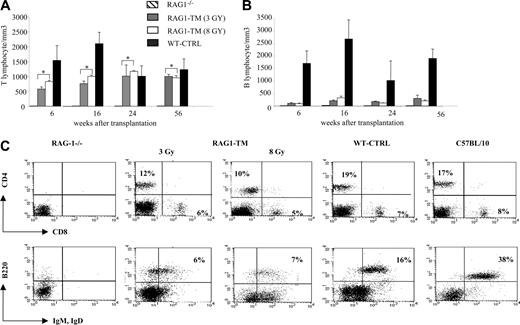
Blood lymphocyte counts after RAG1 retroviral-mediated gene transfer in RAG-1-deficient mice. Peripheral T lymphocytes (A) and B lymphocytes (B) were counted and immunostained at various times. The number of tested mice was as follows: RAG-1-/- (n = 4), RAG1-transduced mice (RAG1-TM, n = 9), and control mice (WT-CTRL, n = 8). Data are expressed as means ± standard deviation (SD). *The comparison of these values is not statistically significant. (C) Blood lymphocyte phenotype after RAG1 retroviral-mediated gene transfer in RAG-1-deficient mice 6 months after treatment. Flow cytometry analysis was performed on RAG-1-/-, RAG1-TM, WT-CTRL, and wild-type mice to detect the proportion of mature T-cell subsets (CD4+CD3+ or CD8+CD3+) and mature B cells (B220+IgM+IgD+). Leukocyte counts were 1000 cells/mm3, 3200 cells/mm3, 3000 cells/mm3, 2400 cells/mm3, and 5000 cells/mm3, respectively, for RAG-1-/-, RAG1-TM (3 Gy), RAG1-TM (8 Gy), WT-CTRL, and wild-type mice. Data are representative of 1 of 9 RAG1-TM mice that gave similar results. Percentages represent the proportion of cells expressing CD4, CD8, B220, IgM, or IgD markers.
Blood lymphocyte counts after RAG1 retroviral-mediated gene transfer in RAG-1-deficient mice. Peripheral T lymphocytes (A) and B lymphocytes (B) were counted and immunostained at various times. The number of tested mice was as follows: RAG-1-/- (n = 4), RAG1-transduced mice (RAG1-TM, n = 9), and control mice (WT-CTRL, n = 8). Data are expressed as means ± standard deviation (SD). *The comparison of these values is not statistically significant. (C) Blood lymphocyte phenotype after RAG1 retroviral-mediated gene transfer in RAG-1-deficient mice 6 months after treatment. Flow cytometry analysis was performed on RAG-1-/-, RAG1-TM, WT-CTRL, and wild-type mice to detect the proportion of mature T-cell subsets (CD4+CD3+ or CD8+CD3+) and mature B cells (B220+IgM+IgD+). Leukocyte counts were 1000 cells/mm3, 3200 cells/mm3, 3000 cells/mm3, 2400 cells/mm3, and 5000 cells/mm3, respectively, for RAG-1-/-, RAG1-TM (3 Gy), RAG1-TM (8 Gy), WT-CTRL, and wild-type mice. Data are representative of 1 of 9 RAG1-TM mice that gave similar results. Percentages represent the proportion of cells expressing CD4, CD8, B220, IgM, or IgD markers.
B- and T-cell characteristics after RAG1 retroviral-mediated gene transfer into RAG-1-deficient mice. (A) Quantitative TCR Vβ and TCR Vα repertoire and (B) representative immunoscope profiles in the spleen cells of RAG1-transduced mice (RAG1-TM) and control mice (WT-CTRL) 6 months after transplantation. The x-axis of immunoscope profiles indicates CDR3 length, and the y-axis displays arbitrary fluorescence intensity of the runoff products. (C) RAG1-TM and WT-CTRL mice were cultured in the presence of anti-CD3+anti-CD28 antibodies. ▪ indicates nonstimulated; ▨, + anti-CD3 + anti-CD28. (D) Total spleen cells were cultured in the presence of LPS. In panels C and D, the proliferative response was measured after addition of [3H]thymidine. Data are expressed as the means ± SD of triplicate wells. ▪ indicates nonstimulated; ▨, + lipopolysaccharide. (E) Immunoglobulins were quantified in the serum of RAG1-TM and WT-CTRL mice at the time of killing or 28 days after immunization with KLH. □ indicates RAG1-TM; ▦, WT-CTRL. (F) Data represent the mean ± SD of 9 RAG1-TM and 8 WT-CTRL mice analyzed. *The comparison of these values is not statistically significant. □ indicates RAG1-TM; ▦, WT-CTRL; and CDR, complementarity-determining region.
B- and T-cell characteristics after RAG1 retroviral-mediated gene transfer into RAG-1-deficient mice. (A) Quantitative TCR Vβ and TCR Vα repertoire and (B) representative immunoscope profiles in the spleen cells of RAG1-transduced mice (RAG1-TM) and control mice (WT-CTRL) 6 months after transplantation. The x-axis of immunoscope profiles indicates CDR3 length, and the y-axis displays arbitrary fluorescence intensity of the runoff products. (C) RAG1-TM and WT-CTRL mice were cultured in the presence of anti-CD3+anti-CD28 antibodies. ▪ indicates nonstimulated; ▨, + anti-CD3 + anti-CD28. (D) Total spleen cells were cultured in the presence of LPS. In panels C and D, the proliferative response was measured after addition of [3H]thymidine. Data are expressed as the means ± SD of triplicate wells. ▪ indicates nonstimulated; ▨, + lipopolysaccharide. (E) Immunoglobulins were quantified in the serum of RAG1-TM and WT-CTRL mice at the time of killing or 28 days after immunization with KLH. □ indicates RAG1-TM; ▦, WT-CTRL. (F) Data represent the mean ± SD of 9 RAG1-TM and 8 WT-CTRL mice analyzed. *The comparison of these values is not statistically significant. □ indicates RAG1-TM; ▦, WT-CTRL; and CDR, complementarity-determining region.
Provirus integration and expression of the hRAG1 transgene
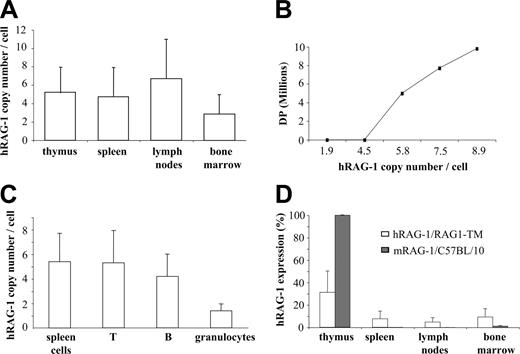
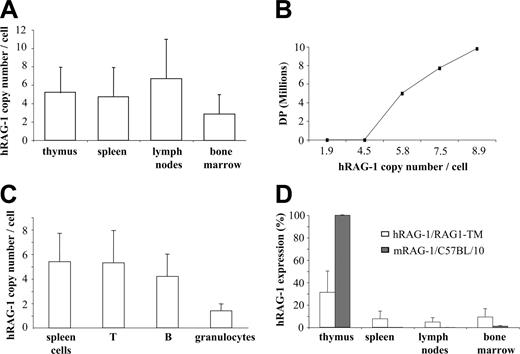
To quantify the integration and expression of the human RAG1 transgene (hRAG1), genomic DNA and RNA were prepared from primary and secondary lymphoid organs as well as from T cells, B cells, and granulocytes sorted from splenocytes. The hRAG1 copy number was found to be higher in lymph node cells, thymocytes, and splenocytes than in the bone marrow cells of RAG1-TM mice (Figure 5A). In addition, a direct correlation was found between the provirus copy number and thymus reconstitution as measured by DP cell counts. In fact, DP cells were not detected when 4 hRAG1 copies per cell or fewer were found in thymocytes whereas the presence of 5 or more hRAG1 copies per cell was associated with a 10-fold increase in DP cells and total thymocyte counts (Figure 5B). These results indicate that in RAG1-TM mice thymic reconstitution occurred only above a threshold of integrated hRAG1 copies. In the spleen, a mean of 5 provirus copies per cell and 1.5 copies per cell was detected in lymphocytes and granulocytes, respectively (Figure 5C). The latter indicates that transduced lymphoid progenitors had a selective advantage over nontransduced cells. Interestingly, the expression of hRAG1 transgene was higher in the thymus than in peripheral lymphoid organs (Figure 5D). However, in the thymus, it corresponded only to a 30% magnitude as compared with mRAG-1 expression in control thymus. Conversely, hRAG-1 expression was found to be slightly more elevated in the bone marrow of RAG1-TM mice as compared with mRAG-1 expression in control mice. This is likely a consequence of the presence of transduced myeloid cells in the former case.
Stability of transgene expression
RAG1-TM mice were analyzed for more than 1 year after treatment. At the last time point (1 year after transplantation), the cell counts of circulating mature T and B cells were identical to those of mice studied 6 months after gene transfer, showing the stability of the immunodeficiency correction over time. To determine whether stable integration of the hRAG1 transgene was achieved in RAG-1-deficient hematopoietic stem cells, 5 × 106 BM cells from 6-month-old reconstituted RAG1-TM mice were injected into RAG-1-/- recipients (n = 10). The mice were analyzed up to 7 months following secondary transplantation. T and B lymphocytes were detectable in all the organs (Figure 6A), although total cell count was low compared with the one observed in primary-treated mice. Immunoscope analysis demonstrated that a diverse TCR repertoire was restored (Figure 6B). Despite low B-cell reconstitution, serum immunoglobulin levels as well as specific antibody responses toward KLH were comparable to control mice (data not shown). A high provirus copy number was also found in the thymus, lymph nodes, and spleen of secondary recipient mice, confirming the requirement for a high RAG1 copy number to achieve reconstitution of T- and B-cell compartments (Figure 6C). A low provirus copy number was found in the bone marrow, likely reflecting a lower proportion of transduced progenitor cells in the animals undergoing secondary transplantation.
Quantification of provirus integration and hRAG1 transgene expression. (A) The hRAG1 copy number was quantified on the DNA extracted from thymus, spleen, lymph nodes, and bone marrow cells (n = 9) 6 months after transplantation. (B) Correlation between hRAG1 copy number and CD4+CD8+ (DP) number in the thymus of RAG1-TM animals 6 months after gene transfer. (C) T cells, B cells, and granulocytes were purified from the spleen before DNA extraction to quantify transgene copy number. Data are expressed as means ± SD. (D) hRAG-1 and mRAG-1 expression were determined on the RNA extracted from thymus, spleen, lymph nodes, and bone marrow cells (n = 9 for RAG1-TM and n = 4 for C57BL/10) 6 months after transplantation. hRAG-1 transcript expression was compared with endogenous acidic ribosomal phosphoprotein expression (PO) and expressed as a percentage of a reference sample (RAG-1 expression in the thymus of a wild-type mouse).
Quantification of provirus integration and hRAG1 transgene expression. (A) The hRAG1 copy number was quantified on the DNA extracted from thymus, spleen, lymph nodes, and bone marrow cells (n = 9) 6 months after transplantation. (B) Correlation between hRAG1 copy number and CD4+CD8+ (DP) number in the thymus of RAG1-TM animals 6 months after gene transfer. (C) T cells, B cells, and granulocytes were purified from the spleen before DNA extraction to quantify transgene copy number. Data are expressed as means ± SD. (D) hRAG-1 and mRAG-1 expression were determined on the RNA extracted from thymus, spleen, lymph nodes, and bone marrow cells (n = 9 for RAG1-TM and n = 4 for C57BL/10) 6 months after transplantation. hRAG-1 transcript expression was compared with endogenous acidic ribosomal phosphoprotein expression (PO) and expressed as a percentage of a reference sample (RAG-1 expression in the thymus of a wild-type mouse).
Toxicity
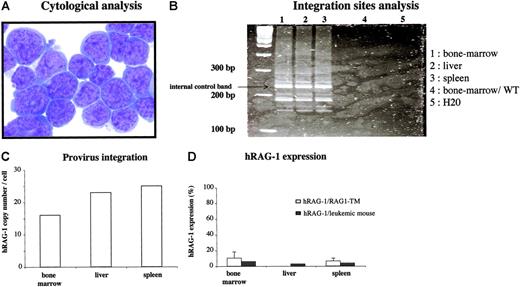
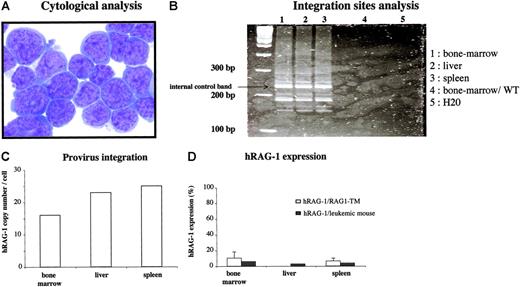
Six months after gene therapy, one mouse from the 8 Gy-irradiated group was killed because of a rapidly arising slow-moving behavior and weight loss. No cells were found in the thymus and lymph nodes of this animal (fewer than 1000 cells) and, conversely, the liver and spleen were enlarged. Of note, the peripheral blood lymphocyte (PBL) analysis performed at 6 and 16 weeks after transplantation did not reveal any aberrant lymphocyte distribution and leukocyte counts (T lymphocytes: 600 cells/mm3 and 1000 cells/mm3 at 6 and 16 weeks, respectively; B lymphocytes: 50 cells/mm3 and 160 cells/mm3 at 6 and 16 weeks, respectively). Hepatosplenomegaly (510 × 106 cells) was the consequence of a monomorphic infiltration of lymphocytes that did not express any tested lymphoid differentiation marker but exhibited the morphology of an undifferentiated acute leukemic proliferation (Figure 7). Provirus integration study showed a very high copy number in the enlarged lymphoid organs (25 copies per cell in the spleen and 23 copies per cell in the liver) as well as in the bone marrow (16 copies per cell). However, hRAG-1 expression is lower than hRAG-1 expression observed in the other RAG1-TM mice. The same integration site profile was observed by LAM-PCR in bone marrow, spleen, and liver, attesting that a clone had preferentially expanded (Figure 7). Of note, murine RAG-2 mRNA expression was detected by RT-PCR in tumor cells, which is strongly suggestive of an early lymphocyte progenitor nature of the tumor cell (Figure S2; see the Supplemental Figures link at the top of the online article, at the Blood website). Finally, no replication-competent retroviruses were detected in the serum of these mice. This observation strongly suggests that this clonal proliferation of undifferentiated cells was the indirect consequence of the high provirus copy number and was not a consequence of an overexpression of hRAG-1. To date, 10 other mice have been engrafted with gene-modified bone marrow cells, and 12 months following gene transfer none of them had developed any lymphoproliferation. Thus, among 30 gene therapy RAG-1-treated mice (9 killed 6 months following gene transfer, 1 lymphoproliferative syndrome 6 months after transplantation, 10 mice undergoing secondary transplantation still alive, and 10 mice undergoing primary transplantation still alive), a single severe adverse effect was observed. Of note, none of the 14 WT-CTRL mice have shown any side effects, excluding the lymphoproliferation likely due to the irradiation step.
Analysis of secondary transplantation in RAG-1-/- deficient mice 6 to 7 months after transplantation. Six months after primary engraftment, mice were killed and 5 × 106 bone marrow cells were injected into secondary RAG-1-/- recipients. (A) Leukocyte cell count was 5000 cells/mm3 in the blood, and total cell counts were 4 × 106 cells in the lymph nodes, 11 × 106 cells in the spleen, and 0.2 × 106 cells in the thymus. The dot plots are representative of 1 mouse of 10 that gave similar results. Percentages represent the proportion of cells expressing CD4, CD8, B220, IgM, or IgD markers. (B) Representative immunoscope profiles of TCR Vβ and TCR Vα analysis in the spleen cells of mice undergoing secondary transplantation 6 months after primary transplantation. The x-axis of immunoscope profiles indicates CDR3 length, and the y-axis displays arbitrary fluorescence intensity of the runoff products. (C) Quantification of provirus integration in RAG-1-/- mice undergoing secondary transplantation. The hRAG1 copy number was determined on the DNA extracted from thymus, spleen, lymph nodes, and bone marrow cells. Data are expressed as means plus or minus SD.
Analysis of secondary transplantation in RAG-1-/- deficient mice 6 to 7 months after transplantation. Six months after primary engraftment, mice were killed and 5 × 106 bone marrow cells were injected into secondary RAG-1-/- recipients. (A) Leukocyte cell count was 5000 cells/mm3 in the blood, and total cell counts were 4 × 106 cells in the lymph nodes, 11 × 106 cells in the spleen, and 0.2 × 106 cells in the thymus. The dot plots are representative of 1 mouse of 10 that gave similar results. Percentages represent the proportion of cells expressing CD4, CD8, B220, IgM, or IgD markers. (B) Representative immunoscope profiles of TCR Vβ and TCR Vα analysis in the spleen cells of mice undergoing secondary transplantation 6 months after primary transplantation. The x-axis of immunoscope profiles indicates CDR3 length, and the y-axis displays arbitrary fluorescence intensity of the runoff products. (C) Quantification of provirus integration in RAG-1-/- mice undergoing secondary transplantation. The hRAG1 copy number was determined on the DNA extracted from thymus, spleen, lymph nodes, and bone marrow cells. Data are expressed as means plus or minus SD.
Discussion
RAG-1-immunodeficient mice were used as a model to demonstrate that ex vivo RAG1 retroviral gene transfer into RAG-1-/- hematopoietic precursor cells can lead to the correction of the immunodeficiency. T and B lymphocytes were detected in RAG1-TM peripheral blood within 4 to 16 weeks after transplantation. Their counts were stable for more than 1 year after gene therapy, attesting to the transduction of pluripotent bone marrow progenitors. In the spleen, blood, lymph nodes, and bone marrow, total lymphocyte counts were identical to those observed in the WT-CTRL animals, whereas in all the organs the B-cell compartment was smaller than in WT-CTRL mice. A tight relationship was observed between the intensity of the conditioning irradiation (3 Gy versus 8 Gy) and thymic reconstitution as DP progenitors were always observed in the thymus of the 8 Gy-irradiated recipient group but in only 40% of the 3 Gy-irradiated recipient group. These data are in line with the observations of Prockop and Petrie28 who recently demonstrated that thymic differentiation is controlled mainly by double negative 3 (DN3) precursor number (CD25+CD44-) located in the intrathymic stromal niches. In the absence of any irradiation, immunodeficient strains of mice with normal or high numbers of DN3 (such as RAG-1-deficient mice) were refractory to thymic reconstitution following HSCT, because of a competition for the occupancy of relevant thymic niches. In contrast, strains of mice with reduced DN3 cell counts (such as IL-7Rα- and γc-deficient mice) were well reconstituted following HSCT. A competition between endogenous DN3 precursors and transduced progenitors probably occurs in the RAG-1-/- recipients. Therefore, 8 Gy irradiation is likely required to eliminate endogenous DN3 cells and facilitate T-cell development from transduced cells. Of note, in the 3 Gy-irradiated group, the percentage of CD4+CD3+ and CD8+CD3+ was not affected by the reduced DP cell counts. This could be explained by intensive expansion of a few DP precursors, as previously shown in a model assessing T-cell differentiation in mice with a limited fraction of competent precursor cells.29 Despite differences in the thymic reconstitution, expansion of transduced cells leads to a nearly normal-sized peripheral T- and B-lymphocyte pool. The T-cell compartment is found to be diverse and functional as shown by Vα and Vβ TCR immunoscope analysis as well as proliferation assays in the presence of mitogens. The mature circulating B lymphocytes were not abundant but functional, as the serum immunoglobulin levels and specific antibody response against the T-cell-dependent KLH antigen were always within normal range. This finding is in accordance with previous observations demonstrating normal serum Ig levels in mice with reduced peripheral B-cell numbers.30 As observed for the thymic reconstitution, we could also make the assumption that endogenous pro-B cells located into the medullar stromal niches disrupt the engraftment of transduced progenitors, thus explaining the low number of mature B cells in the lymphoid organs. It is also possible that the capacity of B-cell precursor expansion is limited as compared with the one of T-cell precursors. Finally, after secondary bone marrow transplantation, lymphocyte development was restored showing that hematopoietic stem cells were successfully transduced.
Characteristics of the severe adverse effect observed in one mouse after RAG1 gene transfer. Six months after primary transplantation, a mouse moving slower and weighing less than its littermates was killed. (A) Cytologic analysis was performed on spleen cells. May Grünwald-Giemsa staining was performed on cytospin slides. Images were visualized using a Leica DM:RB microscope equipped with a 40 ×/0.7 objective lens and a Leica 350F camera (Leica, Wetzlar, Germany). The total original magnification of the image is × 400. (B) LAM-PCR was performed on the DNA extracted from the enlarged organs and on bone marrow cells from a wild-type C57BL/10 mouse. (C) The hRAG1 copy number was quantified on the DNA extracted from the enlarged organs. (D) hRAG-1 expression was determined on the RNA extracted from bone marrow, liver, and spleen cells and compared with the expression in the spleen and bone marrow of the other RAG1-TM mice. hRAG-1 transcript expression was compared with endogenous acidic ribosomal phosphoprotein expression (PO) and expressed as a percentage of a reference sample (RAG-1 expression in the thymus of a wild-type mouse). Data are expressed as means plus or minus SD.
Characteristics of the severe adverse effect observed in one mouse after RAG1 gene transfer. Six months after primary transplantation, a mouse moving slower and weighing less than its littermates was killed. (A) Cytologic analysis was performed on spleen cells. May Grünwald-Giemsa staining was performed on cytospin slides. Images were visualized using a Leica DM:RB microscope equipped with a 40 ×/0.7 objective lens and a Leica 350F camera (Leica, Wetzlar, Germany). The total original magnification of the image is × 400. (B) LAM-PCR was performed on the DNA extracted from the enlarged organs and on bone marrow cells from a wild-type C57BL/10 mouse. (C) The hRAG1 copy number was quantified on the DNA extracted from the enlarged organs. (D) hRAG-1 expression was determined on the RNA extracted from bone marrow, liver, and spleen cells and compared with the expression in the spleen and bone marrow of the other RAG1-TM mice. hRAG-1 transcript expression was compared with endogenous acidic ribosomal phosphoprotein expression (PO) and expressed as a percentage of a reference sample (RAG-1 expression in the thymus of a wild-type mouse). Data are expressed as means plus or minus SD.
It is noteworthy to stress that immunologic reconstitution was only observed when a high copy number of hRAG1 transgene per cell could be detected. A high copy number may be required to compensate for a low transcription rate of hRAG1 transgene, which accounted for only 30% of mRAG-1 expression in control thymus. The slightly higher hRAG-1 expression in the bone marrow of RAG1-TM mice compared with endogenous mRAG-1 expression in the same organ could be explained by the presence of both lymphoid and myeloid transduced cells. Of note, a high transgene copy number may also be required to compensate for a less active RAG-1 protein because human RAG1 cDNA was used. All these data suggest that RAG-1 expression may need to reach a minimum threshold to achieve functionality during lymphoid development. Finally, the requirement for a high copy number could also be explained by the transduction of nonfunctional vector copies due to cryptic splicing or rearrangement of vector sequences prior to integration. These hypotheses were studied by Southern blot and Northern blot analyses of transduced cells and packaging cells, respectively (Figure S1). No rearrangement of vector sequences was detected by Southern blot analysis, as a unique and correct RAG1 transgene band (3.9 kb) was found. Northern blot study revealed the presence of a predominant band at the expected size of the full-length hRAG-1 transcript (6.6 kb). Of note, a minor band of uncorrected size (8 kb) was also detected, but it represented only 10% of the intensity of the bands. These results lead us to conclude that the potential rearrangement of vector sequences probably does not involve the 5′ end of the cDNA where the probes were located. No correlation between hRAG1 copy number and expression was found in the periphery, suggesting that a partial extinction of transgene expression occurred in mature T and B lymphocytes. This hypothesis is reinforced by several studies reporting that MLV-driven transgene expression could be impaired by several mechanisms such as methylation of the proviral LTR.31-33
A selective advantage of RAG1-transduced cells over nontransduced cells was clearly observed, as previously reported in other immunodeficient mouse models of gene therapy9-12 as well as in 2 clinical trials performed for children affected with 2 SCID disease (ie, γc and adenosine deaminase [ADA] deficiencies).5-8 This observation was expected despite the fact that lymphoid differentiation block caused by RAG-1 deficiency occurs at a later stage of hematopoieisis than the one caused by the γc chain or ADA deficiency. However, the selective advantage of RAG1-transduced cells does not appear to be as strong, because a high transduction rate and a cytoreductive treatment were required to achieve the correction of the immunodeficiency. These 2 conditions were not found to be as stringently required for correction of RAG-2 deficiency by gene therapy.11 The discrepancy might be explained in different ways: both therapeutic genes have a different size (1.5 kb versus 3.1 kb for RAG2 and RAG1, respectively) that could influence transgene expression. The genetic background of these knock-out mice is also not identical: C57BL/6 and C57BL/10 for RAG-2 and RAG-1, respectively. As previously described, the phenotype and function of the hematopoietic stem cell could differ according to mouse strain, and this could have an influence on the immune reconstitution.34
The high copy number required to restore the RAG-1 immunodeficiency led to the occurrence of one case of clonal lymphoproliferation among the 30 gene therapy-treated mice. The lymphoproliferation had some characteristics of an acute lymphoblastic leukemia. It was striking that RAG-1 expression was not enhanced but mRAG-2 expression was detected in the tumor cells. This is likely the consequence of the immature lymphocyte progenitor stage of the proliferative cells. One cannot, however, strictly rule out that mRAG-2 expression was triggered by a retroviral insertion into the RAG-1/RAG-2 locus. Such an assumption is reinforced by the fact that no abnormalities were identified in single transgenic mice for RAG1 or RAG2 under the lck promoter whereas RAG1 and RAG2 double transgenic mice present lymphadenopathy and splenomegaly.35 Concordantly with these reports, no evidence of lymphoproliferation could be found in long-term-reconstituted RAG-2 mice with a mean provirus copy number of 2 per cell.11 In the leukemic mouse, the very high copy number of RAG1 provirus was confirmed by Southern Blot analysis (Figure S2) and strengthens the assumption that the leukemic event was caused by insertional mutagenesis. Such an effect was previously reported by Jonkers and Berns,36 who demonstrated that replicative Moloney retroviral vectors integrated in host DNA could activate flanking genes and give rise to clonal or oligoclonal tumors. This was also illustrated by Li et al,37 who first demonstrated that a single vector copy integrated into the murine Evi1 gene is enough to induce activation of this transcription factor. When multiple provirus insertion per cell is detected, the probability of activating genes able to cooperate in the oncogenetic process is greatly enhanced. This is in accordance with the capacity of MLV to become preferentially integrated near the transcription start sites of genes.38,39 The relationship between copy number and clonal proliferation has been recently confirmed with defective retrovirus. Indeed, Modlich et al40 have shown that retroviral gene transfer of multidrug resistance 1 (MDR1) into hematopoietic cells of C57BL/6 mice is associated with the development of leukemia, and the provirus copy number found in leukemic cells is always high (at least 8 copies per cell).
One potential concern of RAG1 gene therapy is the potential toxicity of uncontrolled RAG-1 expression driven by a continuously active promoter. Physiologically, both RAG-1 and RAG-2 proteins need to be expressed to induce a DNA double-strand break at the recombination signal sequences sites,15 and this process is highly and coordinately controlled and restricted to T- and B-cell precursors.41-44 Isolated RAG-1 or RAG-2 protein expression is devoid of activity on DNA, and only the persistent expression of both proteins disturbs thymopoiesis as shown in the double transgenic murine model.35,45 As indeed observed in both RAG1 and RAG2 gene transfer experiences, in the respective RAG-1-/- and RAG-2-/-11 animals, no such toxicity has been observed. Nevertheless, RAG-1-treated animals are currently being monitored for an extended period of 1 year to detect any long-term toxicity of uncontrolled RAG-1 expression.
In conclusion, we have shown that RAG1 gene transfer can correct immunodeficiency associated with RAG-1 defect for more than 1 year. We have also demonstrated that type C retroviral vectors are able to transduce a significant number of murine hematopoietic stem cells as shown by the data of secondary bone marrow transplantation. In this setting, the immunologic reconstitution is only observed when a high provirus copy number is found, and this is associated with an increased risk of toxicity. These results are reminiscent of the adverse effects observed in γc-deficient patients treated by retroviral-mediated γc gene transfer.21 Consequently, lentivirus vectors with self-inactivating LTRs and an internal promoter without any enhancer would be recommended. Lentiviral vectors are expected to more efficiently transduce hematopoietic stem cells,46-48 and this could result in a better T- and B-cell reconstitution as observed in WT-CTRL mice engrafted with wild-type Sca-1 cells. This approach would also be useful to reduce the risk of toxicity related to insertional mutagenesis caused by the LTR enhancer activity, provided that a self-inactivating vector is used.
Prepublished online as Blood First Edition Paper, September 20, 2005; DOI 10.1182/blood-2005-05-2032.
Supported by grants from Inserm, Association Française contre les Myopathies contract GAT0203, Consortium National de Recherche en Génomique (CNRG), European Community (EC) contract QLK3-CT-1999-00859, Consert no. 005242, Inherinet contract QLK3-CT-2001-00427, and Amgen-France. Fellowship (C.L.-P.) is provided by CNRG.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Fabian Gross for technical help, Jérome Maigret for the FACS experiments, Dr Françoise Valensi for the cytologic analysis, and Texcell for the RCR tests.


![Figure 4. B- and T-cell characteristics after RAG1 retroviral-mediated gene transfer into RAG-1-deficient mice. (A) Quantitative TCR Vβ and TCR Vα repertoire and (B) representative immunoscope profiles in the spleen cells of RAG1-transduced mice (RAG1-TM) and control mice (WT-CTRL) 6 months after transplantation. The x-axis of immunoscope profiles indicates CDR3 length, and the y-axis displays arbitrary fluorescence intensity of the runoff products. (C) RAG1-TM and WT-CTRL mice were cultured in the presence of anti-CD3+anti-CD28 antibodies. ▪ indicates nonstimulated; ▨, + anti-CD3 + anti-CD28. (D) Total spleen cells were cultured in the presence of LPS. In panels C and D, the proliferative response was measured after addition of [3H]thymidine. Data are expressed as the means ± SD of triplicate wells. ▪ indicates nonstimulated; ▨, + lipopolysaccharide. (E) Immunoglobulins were quantified in the serum of RAG1-TM and WT-CTRL mice at the time of killing or 28 days after immunization with KLH. □ indicates RAG1-TM; ▦, WT-CTRL. (F) Data represent the mean ± SD of 9 RAG1-TM and 8 WT-CTRL mice analyzed. *The comparison of these values is not statistically significant. □ indicates RAG1-TM; ▦, WT-CTRL; and CDR, complementarity-determining region.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/1/10.1182_blood-2005-05-2032/4/m_zh80010688770004.jpeg?Expires=1770436787&Signature=bBu4qdUlOq2gymWquhYK4ZLLA80zn~ew5I4M-geWdJaWt5sA-F3785B67sYY9yDCTWgtui~1UDYt-bpH84YvERN7Mm7~y0ugwk7urNiU33SkZK5xnwj6UoqbjxEVqU0TQI4PRaeadt7hkbI7azEN0rwWO4qORahGRpCTvMiUrV4eqpitaWsykmeVhDz7Co0YaypWcFQ4mE1q30HdxXICKAYMynih3IHUt7D8I3yMD7Gx93VEyUY75Q5BHlBPGXKEkx4KzdYoI7CWrap0YT8yXagxcJJZ3Ih-bmvHcWvP8ScBwgDpqqoTcU7PlJph4fFcoRhr51lfscnE6epUc3SmiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 4. B- and T-cell characteristics after RAG1 retroviral-mediated gene transfer into RAG-1-deficient mice. (A) Quantitative TCR Vβ and TCR Vα repertoire and (B) representative immunoscope profiles in the spleen cells of RAG1-transduced mice (RAG1-TM) and control mice (WT-CTRL) 6 months after transplantation. The x-axis of immunoscope profiles indicates CDR3 length, and the y-axis displays arbitrary fluorescence intensity of the runoff products. (C) RAG1-TM and WT-CTRL mice were cultured in the presence of anti-CD3+anti-CD28 antibodies. ▪ indicates nonstimulated; ▨, + anti-CD3 + anti-CD28. (D) Total spleen cells were cultured in the presence of LPS. In panels C and D, the proliferative response was measured after addition of [3H]thymidine. Data are expressed as the means ± SD of triplicate wells. ▪ indicates nonstimulated; ▨, + lipopolysaccharide. (E) Immunoglobulins were quantified in the serum of RAG1-TM and WT-CTRL mice at the time of killing or 28 days after immunization with KLH. □ indicates RAG1-TM; ▦, WT-CTRL. (F) Data represent the mean ± SD of 9 RAG1-TM and 8 WT-CTRL mice analyzed. *The comparison of these values is not statistically significant. □ indicates RAG1-TM; ▦, WT-CTRL; and CDR, complementarity-determining region.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/1/10.1182_blood-2005-05-2032/4/m_zh80010688770004.jpeg?Expires=1770447027&Signature=uOAqjG7a3xLXzXKKlKWfmSxEgSu1pYhyh9ky5R48fGzFmiQ0vrsbfHsPDQ4cSKFqm9FdKPLiA0SgzgUAZrWR21-lC4b2F~QKtd6QydD7UFgAqLFpVTkD36~e5tM35D-Mt~J2M3cwrnBK--vv5FqcAT3hsVnbaYZAUCIq7W~fi5-OQw8EPnfPsflamvCBuSjFhtntFcqeKX4-zCfaaDrU7YAr82uKAQjAk-JUoxFSivFkGwGQraH6ffYDL5sovb~4oJk8UduwyVzHQq-pJTzHhyPYzIGiFHcKrXLFkZovz-ItjMZd9nm4MScc6wksip5~1dChdISowiDKZyexvqQdsA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)