Comment on Libura et al, page 2124
Nonrandom chromosomal translocations are considered to be causal events leading to leukemic transformation. However, the mechanism(s) that cause these translocations are poorly understood. Libura and colleagues report exciting new findings on the mechanism(s) that lead to chromosomal translocation.
A novel variant of the “Pottery Barn rule” was invoked during the recent US presidential election. This rule (“you break it, you fix it”) also applies to the repair of mammalian DNA. It has been estimated that approximately 50 DNA double-strand breaks occur during S phase of a normal cell division cycle, and additional breaks may occur during other phases of the cell cycle. Moreover, a large number of environmental insults, including genotoxic cancer chemotherapy, can also induce DNA double-strand breaks. In general, there are 2 mechanisms available for repair of these breaks in mammalian cells, homologous recombination (HR) and nonhomologous end joining (NHEJ). HR uses a homologous DNA strand as a template for repair, and can be regarded as error free. NHEJ repairs DNA double-strand breaks by joining together 2 nonhomologous DNA segments. Since this repair is imperfect, NHEJ potentially compromises genetic information. NHEJ is characterized by short deletions, indirect or direct repeats, and short (< 10 bp) regions of microhomology at the double-strand break junction. Analysis of chromosomal translocation breakpoint junctions has revealed many of the hallmarks of NHEJ, such as inverted repeats and microhomology,1 suggesting that an “errorprone” NHEJ process was responsible for the translocation.FIG1
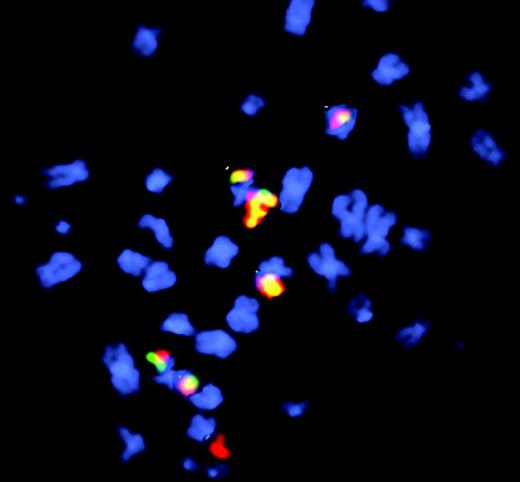
Fluorescence in situ hybridization to visualize etoposide-induced chr11 rearrangements in cells after long recovery period. See the complete figure in the article beginning on page 2124.
Fluorescence in situ hybridization to visualize etoposide-induced chr11 rearrangements in cells after long recovery period. See the complete figure in the article beginning on page 2124.
More than a decade ago,2 acute myeloid leukemia was noted to develop as a second malignancy in patients treated for a primary malignancy with topoisomerase II poisons. These patients often had translocations involving the MLL gene located on chromosome 11q23. Subsequent studies revealed that many of these MLL translocations occurred within a 1.8-kb “hotspot” that contained a site sensitive to DNA cleavage in vivo following treatment with topoisomerase II poisons.3,4 This site mapped to a scaffold/matrix attachment region (S/MAR), and was also hypersensitive to cleavage by DNAse I and apoptotic nucleases, suggesting that it may be a region prone to DNA double-strand breaks.3,5 In this issue, Libura and colleagues identify imperfect DNA repair and apparent chromosomal translocations within this region in human CD34 cells following treatment with the topoisomerase II poison etoposide in vitro. The authors used an inverse polymerase chain reaction (IPCR) approach6 to clone these breakpoint junctions. The authors found that after a brief recovery period, almost half of the IPCR clones recovered represented apparent MLL chromosomal translocations that contained MLL sequences fused to distant genomic regions, but after a longer recovery period, only a single clone represented an apparent translocation. The authors cautiously refer to clones that contain non-MLL sequences fused to MLL as apparent translocations, because they may in fact be insertions of large (> 1 kb) segments of DNA rather than chromosomal translocations. The observation that the proportion of clones with apparent translocations decreased dramatically with longer recovery times suggests that the majority of clones with apparent translocations were unable to proliferate. As noted by the authors, this report now provides a platform upon which to build future studies, such as the effect of defective DNA repair proteins on MLL fusions following genotoxic cancer chemotherapy. ▪