Abstract
Divalent metal transporter 1 (DMT1) is a transmembrane protein crucial for duodenal iron absorption and erythroid iron transport. DMT1 function has been elucidated largely in studies of the mk mouse and the Belgrade rat, which have an identical single nucleotide mutation of this gene that affects protein processing, stability, and function. These animals exhibit hypochromic microcytic anemia due to impaired intestinal iron absorption, and defective iron utilization in red cell precursors. We report here the first human mutation of DMT1 identified in a female with severe hypochromic microcytic anemia and iron overload. This homozygous mutation in the ultimate nucleotide of exon 12 codes for a conservative E399D amino acid substitution; however, its pre-dominant effect is preferential skipping of exon 12 during processing of pre–messenger RNA (mRNA). The lack of full-length mRNA would predict deficient iron absorption in the intestine and deficient iron utilization in erythroid precursors; however, unlike the animal models of DMT1 mutation, the patient is iron overloaded. This does not appear to be due to up-regulation of total DMT1 mRNA. DMT1 protein is easily detectable by immunoblotting in the patient's duodenum, but it is unclear whether the protein is properly processed or targeted.
Introduction
The last 5 years have been marked by an explosion of knowledge in the field of iron metabolism. These discoveries include identification of the proteins involved in iron absorption and trafficking; discovery of a small peptide, hepcidin, which appears to regulate iron uptake; and elucidation of the role of iron in gene expression. One of the newly identified proteins is divalent metal transporter 1 (DMT1), which is expressed at the brush border of enterocytes in the proximal duodenum where it is presumed to mediate pH-dependent uptake of ferrous iron from the gut lumen.1,2 In the erythroblast, the protein is present in the endosomal membrane, where it appears to function in transport of iron released from the transferrin-receptor complex toward the mitochondria, site of heme synthesis,3 via a pathway that remains to be defined. In rodent models, mutation of a single nucleotide results in substitution of Arg for Gly at position 185 (G185R) of the DMT1 gene and leads to hypochromic/microcytic anemia and iron deficiency.4,5
The predicted structure of the protein includes 12 transmembrane domains, asparagine-linked glycosylation signals in an extracytoplasmic loop, membrane targeting motifs, and a consensus transport motif that is common to homologous cation transport proteins found in other species.6,7 Four major mammalian DMT1 isoforms, resulting from alternative splicing at the 5′ and 3′ ends of pre–messenger RNA (mRNA) are known.8 Isoform I has an iron responsive element (IRE) in the 3′ untranslated region, whereas isoform II lacks the IRE, and the C-terminal 18 amino acids are replaced by a novel 25–amino acid segment.6 The duodenum expresses primarily isoform I, whereas erythroblasts express chiefly isoform II; other tissues such as the kidney, brain, liver, and thymus express both isoforms.9 Alternative 5′ exons designated 1A and 1B have also been described; these messages result from different promoters and transcription start sites with mutually exclusive splicing of the alternative first exons to exon 2.8 The exon 1A isoform appears to insert an additional 29 to 31 amino acids onto the N-terminus of the protein. Skipping of exons 10 and 12 during processing of DMT1 mRNA has also been reported, but its significance in vivo has not yet been explored.6
Previously, we reported a young woman with hypochromic/microcytic anemia and iron overload due to a defect in iron transport and utilization in erythroid cells.10 Here, we describe that the defect is caused by an exonic mutation in the DMT1 gene. This mutation exaggerates skipping of exon 12, and results in a patient phenotype that differs from that of the rodent model.
Patients, materials, and methods
All of the propositus' tissues, and those of her sister, were obtained with the approval of the institutional review board (IRB) committee of Palacky University in Olomouc, Czech Republic. The tissues of the control subjects were obtained with the approval of the IRB committee of Baylor College of Medicine; control duodenal samples were generously donated by the tissue procurement Core Laboratory of the Gastrointestinal Center of the Baylor College of Medicine. Informed consent for all subjects was provided according to the Declaration of Helsinki.
Exclusion of thalassemia
Thalassemia in the propositus was excluded in the laboratory of the late Dr Huisman, Augusta, GA.11 The patient had a normal level of HbA2 (1.5% by cation exchange high-performance liquid chromatography [HPLC]) and slightly elevated levels of HbF (2.4% by alkali denaturation and HPLC; gamma chain composition of 64% G gamma and 36% A gamma by reverse-phase HPLC). Abnormal hemoglobins were excluded by protein electrophoresis and HPLC. No inclusion bodies of precipitated globin chains were detected in the patient's erythroid cells after brilliant cresyl blue staining. Deletional types of α and δ-β thalassemias were excluded using Southern blot analysis of the α and β globin gene loci. In addition, the β globin gene, including the 5′ promoter and 3′ untranslated regions (UTRs), was sequenced and no mutation was found.
Preparation of peripheral and in vitro–cultured hematopoietic cells
Platelets, lymphocytes, and granulocytes were separated by differential centrifugation and isopyknic density gradient separation using standard protocols.12 Granulocyte-macrophage–colony-forming unit (CFU-GM) colonies were cultured in clonogenic cultures in 35-mm Petri dishes by using semisolid medium, maintained in a humidified atmosphere at 5% CO2 and 21% O2 at 37°C and isolated using the standard protocols.13 Epstein-Barr virus (EBV)–transformed lymphocytes were prepared and maintained, and their RNA was isolated as previously described.14
Screening for Tfr1 mutations
Single-stranded conformation polymorphism (SSCP) analysis was performed to detect mutations in the coding regions of the transferrin receptor 1 gene. For the patient and the 3 healthy controls, each of the 18 exons with their intron/exon boundaries was amplified by polymerase chain reaction (PCR) using specific primers and 32P-labeled deoxycytidine 5′-triphosphate (dCTP). Two microliters of 32P-labeled product mixed with 2 μL sequencing loading dye was heated for 3 minutes at 95°C to denature, and then it was snap-cooled on ice. The samples were then loaded and run on a nondenaturing sequencing 0.5x mutation detection enhancement (MDE) polyacrylamide gel (Biowhittaker Molecular Applications, Rockland, ME) with constant power (6 W) at room temperature for 16 hours. After electrophoresis, the gel was dried for one hour at 80°C and exposed to an x-ray film at 80°C for autoradiography.15 This approach worked well for all but the 12th exon, which was sequenced directly as described for DMT1 below. The expression of transferrin receptor 1 protein was determined by immunofluorescence analysis with commercially available Tfr antibody (CD71; Santa Cruz Biotechnology, Santa Cruz, CA).
Reverse transcription–PCR amplification of the human DMT1 and ferroportin sequences
Total RNA was prepared from cells and tissues of healthy controls and the patient using the Trizol reagent (Life Technologies, Grand Island, NY) following the manufacturer's instructions. Total RNA was reverse transcribed in a 20 μL reaction using the reverse transcription (RT)–PCR kit (Stratagene, La Jolla, CA) with random primers according to the manufacturer's instructions. Amplification of DMT1 cDNA was performed using 4 sets of primers encompassing all coding exons. Segment 1 (exons 2-5) forward primer: 5′-TCTAAGAACTCAGCCACTCAGG-3′, reverse primer: 5′-GACCTTGGGATACTGACGGTGA-3′; segment 2 (exons 5-9) forward primer: 5′-GCAGCTAGACTGGGAGTGGTTAC-3′, reverse primer: 5′-ACTTGACTAAGGCAGAATGCAG-3′; segment 3 (exons 9-13) forward primer: 5′-GCATCGTGGGAGCTGTCATCAT-3′, reverse primer: 5′-GGATGATGGCAATAGAGCGAGT-3′; segment 4 (exons 13-16) forward primer: 5′-CCTGAACCTAAAGTGGTCACGC-3′, reverse primer: 5′-CTCTGATGGCTACCTGCAGAAG-3′. Depending on the tissue, 2 μLto 5 μL cDNA was used in each PCR reaction, 1x PCR buffer, 2 mM dNTPs, 2 U to 5 U Taq polymerase in a 50 μL reaction mixture. After initial denaturation at 95°C for 5 minutes, amplification was performed for 30 cycles for 95°C for 30 seconds, 63°C for 30 seconds, and 72°C for 30 seconds, then 72°C for 10 minutes. The PCR product was analyzed on a 1% agarose gel then purified on a QIAquick PCR purification column (Qiagen, Valencia, CA). Sequencing was performed by Seqwright (Houston, TX) using a fluorescent-tagged dideoxy chain termination method with an ABI automated sequencer (Applied Biosystems, Foster City, CA). Sequencing of the coding regions of the ferroportin gene was accomplished in a similar manner to that described for DMT1.
Real-time PCR
cDNA derived from reticulocytes and duodenal biopsy samples as described in the previous section was subjected to real-time quantitative PCR in an ABI Prism 7700 Sequence Detection System (Applied Biosystems). Total DMT1 transcript was determined by use of a commercially available kit with primers and detection probe near the exon 15/exon 16 junction (Applied Biosystems). Full-length DMT1 transcript was determined by use of a system with forward primer in exon 11 (5′-GACACTGGCTGTGGACATCTAC-3′), reverse primer in exon 12 (5′-CAGCAGGCCCAAAGTAACATC-3′), and detection probe at the exon 11/12 junction (5′-CAGCACAACACCCCCTTT-3′ labeled with the FAM fluorophore). All results were normalized to the amount of 18S rRNA in the samples as previously described.15 The results are expressed relative to a calibrator, which was designated as the sample with the lowest expression level of DMT1 within each assay. For example, within each assay ΔCT for a particular sample was calculated as: (threshold cycle for that sample + dilution factor) - threshold cycle for 18S rRNA for that sample. ΔΔCT was calculated as the difference between the ΔCT for a particular sample and the ΔCT of a calibrator. Relative expression of each particular transcript is then calculated as 2- ΔΔCT. Each sample was examined in duplicate and the data are expressed as a range by evaluating the expression 2- ΔΔCT with ΔΔCT +s and ΔΔCT - s, where s is the standard deviation of the ΔΔCT value.
Analysis for DMT1 1285 G gC mutation
Using primers specific for the exon/intron boundary of exon 12/intron 12, genomic DNA from 108 healthy subjects, the propositus, and the propositus' sister was subjected to PCR amplification (432 base pair [bp]). PCR products were then digested with BsiW I endonuclease (New England Biolabs, Beverly, MA) as per the manufacturer's instructions. Wild-type PCR product has no predicted cleavage site for this enzyme; cleavage of the mutant 1285 G > C PCR fragment generated 2 smaller fragments with 291 bp and 141 bp. Cleavage products were evaluated on 1% agarose gels.
Immunoblotting
Total duodenal protein was isolated from the phenol-ethanol supernatant of the Trizol extraction as described by the manufacturer (Life Technologies) except that only a single extraction with 0.3 M guanidine hydrochloride in 95% ethanol was performed. Protein pellets were solubilized in Laemmli buffer and the protein concentration was determined by Bradford assay. Protein extracts were incubated for 20 minutes at room temperature before sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 10% polyacrylamide) and transfer to polyvinylidene difluoride (PVDF) membranes. PVDF membranes were preincubated with blocking solution (0.2% skim milk in Tris-buffered saline [TBS]) at room temperature for 60 minutes, washed with TTBS (0.1% Tween 20 in TBS), then incubated overnight with primary antibodies at 4°C. For immunoblotting, primary antibodies were used as follows: rabbit anti-Nramp2 NT (1/200; a generous gift of Dr Philippe Gros, McGill University, Montreal, QC, Canada), rabbit anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1/2000; Abcam, Cambridge, MA). After incubation with primary antibody, the blots were washed with TTBS, then incubated with alkaline phosphatase–labeled anti-rabbit secondary antibody (1/3000) at room temperature for 90 minutes, and the signal was visualized with Immun-Star (Biorad, Hercules, CA).
Results
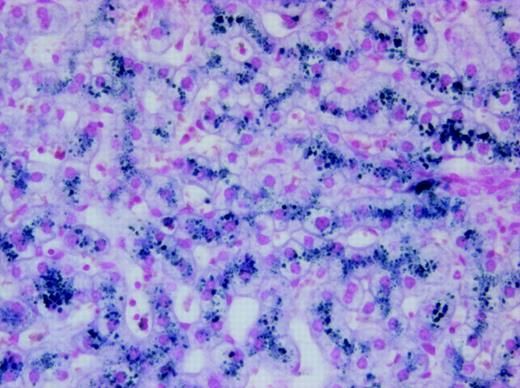
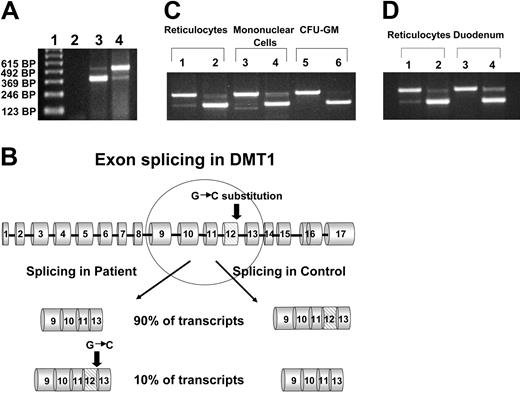
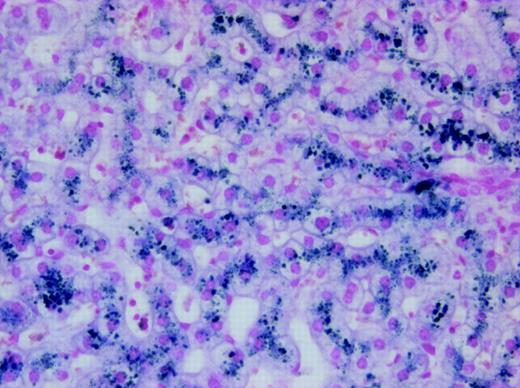
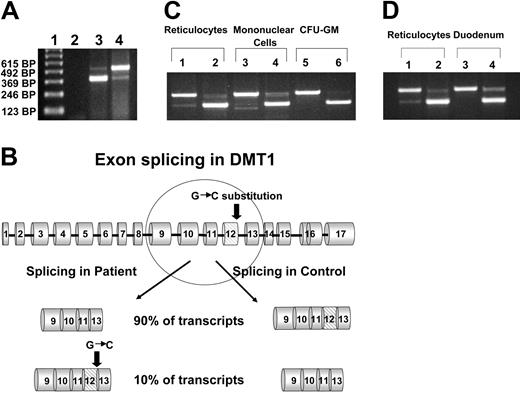
We identified a female, the result of a consanguineous union, who came to medical attention at the age of 3 months due to severe hypochromic, microcytic anemia. Bone marrow exam revealed erythroid hyperplasia with features of abnormal erythroid maturation; reticulocyte count was normal to slightly elevated. No evidence of thalassemia was found. Serum iron levels have been consistently elevated (35 μM-47 μM; normal values 14.5 μM-26.0 μM), ferritin levels have been normal to slightly increased (58 ng/mL-305 ng/mL; normal values 20 ng/mL-150 ng/mL), and serum transferrin receptor levels have remained highly increased. The patient has received, on average, one transfusion per year, but her development has been unremarkable with no obvious organ dysfunction.15 At the age of 8 years, she developed mild liver function abnormalities, and liver biopsy at age 19 demonstrated significantly increased iron deposition in both Kupffer cells and hepatocytes (Figure 1) despite the fact that the volume of transfusions she had received was not sufficient for development of hepatic hemosiderosis. Culture of her erythroid progenitors revealed small, poorly hemoglobinized cells. However, this defect was corrected by salicylaldehyde isonicotinoyl hydrazone saturated with iron (FeSIH), an agent that transfers iron independently of DMT1 and the transferrin receptor.10 We hypothesized that this erythroid defect was due to defective iron utilization in the erythroid precursors. Transferrin receptor 1 (Tfr1) expression in the patient's reticulocytes was normal and analysis of the Tfr1 demonstrated no mutation of the coding regions or intron/exon boundaries. Similarly, sequencing of the coding region of ferroportin cDNA showed no mutation. Analysis of the reticulocyte DMT1 mRNA sequence was accomplished by amplifying total cDNA with 4 separate sets of primers, which span all coding exons. Examination of the exon 9 to 13 region of the DMT1 mRNA in the normal reticulocytes revealed 2 transcripts, one larger band comprising about 90% of the total and a second smaller band comprising the remaining 10%. By contrast, the patient's reticulocytes revealed an inverse ratio of the larger and smaller bands (Figure 2A). Sequencing of the larger and smaller transcripts from both the patient and control subject revealed that the larger transcript contained the full-size exon 9-13 message, while the smaller transcript had exon 12 (central coding region of DMT1) deleted. The larger transcript from the patient, however, had a homozygous G>C missense mutation of the ultimate nucleotide of exon 12 (DMT1 1285G>C) that changes Glu 399 to Asp. Evaluation of the patient's genomic DNA confirmed the exon 12 mutation and revealed no mutations in introns 11, 12, or 13 or in the 3′ UTR encoding the IRE or its vicinity. Analysis of 108 healthy subjects found none with DMT1 1285G>C. A schematic outline of the DMT1 gene mutation and alternate pre-mRNA processing is depicted in Figure 2B.
Photomicrograph of liver biopsy from the patient showing iron deposition in both hepatocytes and Kupffer cells. Staining technique: Perl Prussian blue stain. Original magnification, × 200. The image was acquired using an Olympus BX50 microscope equipped with a UPlan F1 × 20/0.5 NA objective. The photography was performed using an Olympus DP50 digital camera and the Viewfinder Lite version 1.0 program (Pixera, Los Gatos, CA).
Photomicrograph of liver biopsy from the patient showing iron deposition in both hepatocytes and Kupffer cells. Staining technique: Perl Prussian blue stain. Original magnification, × 200. The image was acquired using an Olympus BX50 microscope equipped with a UPlan F1 × 20/0.5 NA objective. The photography was performed using an Olympus DP50 digital camera and the Viewfinder Lite version 1.0 program (Pixera, Los Gatos, CA).
Amplification of the exon 9 to 13 region of DMT1 mRNA from reticulocytes. (A) RNA from the patient's reticulocytes and those of a healthy control was isolated, and total cDNA was synthesized as described in “Patients, materials, and methods.” The exon 9 to 13 region of DMT1 cDNA was amplified by PCR and the PCR products were separated on a 1% agarose gel. Lane 1, 123-bp ladder; lane 2, no template control; lane 3, patient's reticulocytes; lane 4, healthy control's reticulocytes. The upper band in lanes 3 and 4 corresponds to a size of 492 bp, the lower band in lanes 3 and 4 corresponds to a size of 372 bp. (B) Schematic representation of DMT1 pre-mRNA processing in the patient and in wild-type control. (C) Amplification of the exon 9 to 13 region of DMT1 mRNA from various hematopoietic cells. RNA from the patient's reticulocytes, mononuclear cells, and CFU-GMs, and from corresponding cells from a healthy control was isolated and total cDNA was synthesized. The exon 9 to 13 region of DMT1 cDNA was amplified by PCR, and the PCR products were separated on a 1% agarose gel. Lanes 1, 3, and 5 are from a healthy control; lanes 2, 4, and 6 are from the patient. Platelets and granulocytes from the patient and healthy controls were also examined and data were similar to mononuclear cells (data not shown). (D) Amplification of the exon 9 to 13 region of DMT1 mRNA from reticulocytes and duodenum. RNA was isolated from a small sample of the patient's duodenum (obtained at endoscopy) and from the duodenum of a healthy control, and total cDNA was synthesized. The exon 9 to 13 region of DMT1 cDNA was amplified by PCR, and the PCR products were separated on a 1% agarose gel. Lanes 1 and 3 are from a healthy control; lanes 2 and 4 are from the patient.
Amplification of the exon 9 to 13 region of DMT1 mRNA from reticulocytes. (A) RNA from the patient's reticulocytes and those of a healthy control was isolated, and total cDNA was synthesized as described in “Patients, materials, and methods.” The exon 9 to 13 region of DMT1 cDNA was amplified by PCR and the PCR products were separated on a 1% agarose gel. Lane 1, 123-bp ladder; lane 2, no template control; lane 3, patient's reticulocytes; lane 4, healthy control's reticulocytes. The upper band in lanes 3 and 4 corresponds to a size of 492 bp, the lower band in lanes 3 and 4 corresponds to a size of 372 bp. (B) Schematic representation of DMT1 pre-mRNA processing in the patient and in wild-type control. (C) Amplification of the exon 9 to 13 region of DMT1 mRNA from various hematopoietic cells. RNA from the patient's reticulocytes, mononuclear cells, and CFU-GMs, and from corresponding cells from a healthy control was isolated and total cDNA was synthesized. The exon 9 to 13 region of DMT1 cDNA was amplified by PCR, and the PCR products were separated on a 1% agarose gel. Lanes 1, 3, and 5 are from a healthy control; lanes 2, 4, and 6 are from the patient. Platelets and granulocytes from the patient and healthy controls were also examined and data were similar to mononuclear cells (data not shown). (D) Amplification of the exon 9 to 13 region of DMT1 mRNA from reticulocytes and duodenum. RNA was isolated from a small sample of the patient's duodenum (obtained at endoscopy) and from the duodenum of a healthy control, and total cDNA was synthesized. The exon 9 to 13 region of DMT1 cDNA was amplified by PCR, and the PCR products were separated on a 1% agarose gel. Lanes 1 and 3 are from a healthy control; lanes 2 and 4 are from the patient.
We also examined platelets, granulocytes, monocytes, and granulocytic progenitors (CFU-GMs) grown in vitro from the patient and a healthy control (Figure 2C). Platelets, granulocytes, and monocytes in healthy individuals contained, primarily, the full-size transcript and 2 smaller bands, one of which was an exon 12–skipped transcript and the other an exon 10–skipped transcript (confirmed by sequencing). In contrast, the equivalent cell types from the patient showed, primarily, an exon 12–skipped message; exon 10–skipped cDNA was not detectable in the patient. As in reticulocytes, DMT1 mRNA from the patient's CFU-GMs demonstrated, primarily, a smaller message with exon 12 skipped. Cultured CFU-GMs from a healthy control showed 100% of the large transcript. Interestingly, EBV lymphocytes from healthy subjects also had only detectable, full-size DMT1 mRNA; similar cells were not available from the propositus. The presence of exon skipping did not appear to be isoform dependent, as EBV lymphocytes expressed approximately equal amounts of mRNA isoforms I and II, but did not demonstrate exon skipping (data not shown).
In order to determine whether exon skipping was limited to hematopoietic cells, we obtained duodenum from healthy subjects and from the patient. Evaluation of DMT1 mRNA from the duodenum of 5 control subjects (2 with Fe deficiency) showed no evidence of exon 12 skipping. Evaluation of the exon 9 to 13 regions of DMT1 mRNA isolated from the patient's duodenal biopsy again demonstrated the presence of 2 transcripts (Figure 2D). Quantitation of the relative amounts of total DMT1 transcript in the duodenum from the patient and from healthy controls by real-time PCR revealed no significant differences in total DMT1 transcript (Table 1). Using a probe that was specific for full-length (exon 12–containing) DMT1 transcript demonstrated that in the duodenum, the patient had 2- to 40-fold less full-length transcript than controls. The largest differences were between the patient and controls 2 and 4, who were the iron-deficient subjects. Similar studies were also performed to compare total and full-length DMT1 mRNA levels in reticulocytes from the patient and 3 healthy controls. Again, total DMT1 mRNA levels in patient reticulocytes fell within the range of the healthy controls; however, the patient reticulocytes contained 14- to 28-fold less full-length DMT1 transcripts than healthy controls (Table 2).
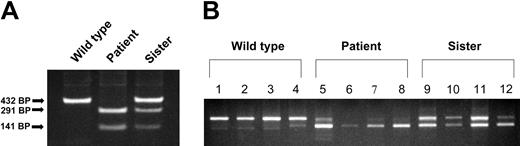
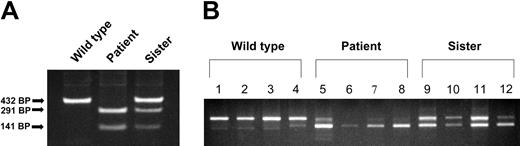
A blood sample was also obtained from the patient's asymptomatic sister (parents declined to participate) from which reticulocytes, monocytes, granulocytes, and platelets were isolated. Analysis of the genomic DNA from the patient, her sister, and a healthy control by PCR amplification of the exon 12/intron 12 boundary followed by endonuclease digestion revealed that the sister was a heterozygote for the mutation (Figure 3A). This result was confirmed by analysis of the exon 9 to 13 region of the DMT1 mRNA in the sister's granulocytes, monocytes, and platelets, revealing approximately equal amounts of the skipped and non-skipped transcripts (Figure 3B). However, her reticulocytes had a relatively greater proportion of a smaller message with exon 12 skipped (based on scanning densitometry of the gel, ratio of full-length to exon 12–skipped transcript was 7:3 in the sister versus 9:1 in the patient).
Comparison of exon 12–skipping in various tissues from wild-type, patient, and patient's sister (heterozygote). (A) Genomic DNA was isolated from reticulocytes and the exon/intron boundary of exon 12/intron 12, and was amplified by PCR. PCR products were subjected to digestion with BsiW I endonuclease and then analyzed on a 1% agarose gel. (B) RNA was isolated from mononuclear cells, granulocytes, platelets, and reticulocytes, and total cDNA was synthesized. The exon 9 to 13 region of DMT1 cDNA was amplified by PCR and the PCR products were separated on a 1% agarose gel. Lanes 1, 5, and 9 are from mononuclear cells; lanes 2, 6, and 10 are from granulocytes; lanes 3, 7, and 11 are from platelets; lanes 4, 8, and 12 are from reticulocytes.
Comparison of exon 12–skipping in various tissues from wild-type, patient, and patient's sister (heterozygote). (A) Genomic DNA was isolated from reticulocytes and the exon/intron boundary of exon 12/intron 12, and was amplified by PCR. PCR products were subjected to digestion with BsiW I endonuclease and then analyzed on a 1% agarose gel. (B) RNA was isolated from mononuclear cells, granulocytes, platelets, and reticulocytes, and total cDNA was synthesized. The exon 9 to 13 region of DMT1 cDNA was amplified by PCR and the PCR products were separated on a 1% agarose gel. Lanes 1, 5, and 9 are from mononuclear cells; lanes 2, 6, and 10 are from granulocytes; lanes 3, 7, and 11 are from platelets; lanes 4, 8, and 12 are from reticulocytes.
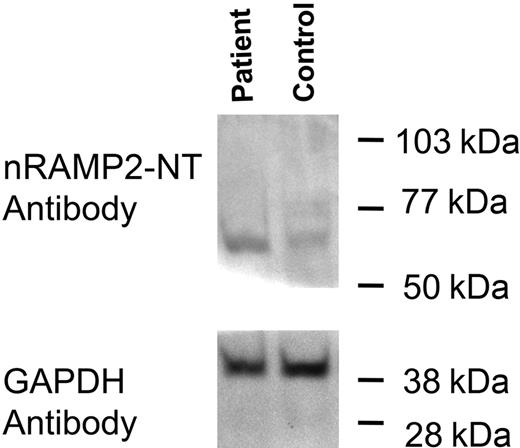
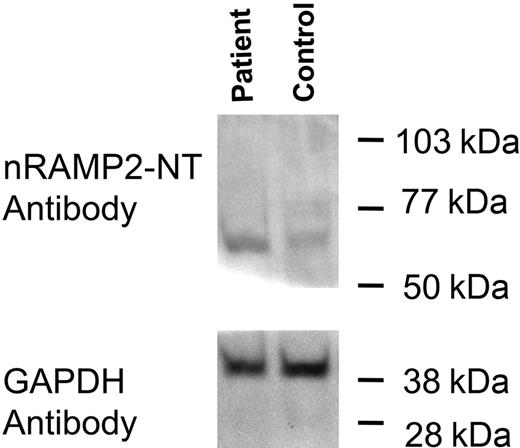
Total duodenal protein was extracted from the lower phase of the trizol extract of duodenum and used for Western blotting. Immunoblotting studies of the patient's sample along with a control sample of duodenum showed a single band with a molecular weight of approximately 65 kDa (Figure 4). This band corresponded to a band seen in the protein extract of Caco-2 cells (data not shown) and to the molecular weight of DMT1 in Caco-2 cells reported previously.16 This study suggests that the DMT1 protein level in the patient is at least equal to and possibly greater than that seen in the healthy control. Similar studies of the patient's reticulocytes were precluded by a limited amount of material.
Detection of DMT1 protein in duodenum. Total duodenal extract was prepared as described: 22 μg total protein was loaded into each lane and separated by SDS-PAGE on a 10% acrylamide gel followed by transfer to PVDF membrane. Membranes were cut in half just below the 50-kDa standard; the upper half of the blot was incubated with the nRAMP2-NT antibody, whereas the lower half was incubated with the GAPDH antibody as an internal loading control.
Detection of DMT1 protein in duodenum. Total duodenal extract was prepared as described: 22 μg total protein was loaded into each lane and separated by SDS-PAGE on a 10% acrylamide gel followed by transfer to PVDF membrane. Membranes were cut in half just below the 50-kDa standard; the upper half of the blot was incubated with the nRAMP2-NT antibody, whereas the lower half was incubated with the GAPDH antibody as an internal loading control.
Discussion
We report the first human mutation of the DMT1 gene. In our patient, a single nucleotide substitution predicts a peptide sequence with a conservative amino acid substitution. However, substitution of this nucleotide leads to a gross exaggeration of the normal low level of exon 12 skipping during processing of the DMT1 mRNA present in erythroid cells. Interestingly, exon 12 skipping was also observed in patient tissues such as duodenum and nonerythroid blood cells, but not in the EBV transformed lymphocytes, CFU-GMs, or duodenum of healthy controls. These data raise several points about DMT1 protein. First, although the alternatively processed DMT1 mRNA retains its reading frame, it is not yet known whether the shorter mRNA results in synthesis of a functional protein product. Deletion of this exon removes transmembrane domain 8, which would be predicted to interfere with correct protein insertion into the membrane. Interestingly, we were able to detect DMT1 protein in the patient's duodenal sample in a magnitude at least comparable to, if not greater than, that in the control duodenum sample; however, it is not clear whether the translational product of exon 12–skipped mRNA is present or whether our antibody and detection assay can identify it. Additionally, our results do not address intracellular targeting of the protein. In rats, Trinder et al17 have reported that DMT1 is located primarily in the apical membrane of the villus enterocytes in iron-deficient animals, but mainly in the cytoplasm of iron-loaded animals. Thus, if a mutant protein is synthesized, it will be important to determine if it reaches its proper destination in the enterocytes and or reticulocytes. This is emphasized by the recent work of Touret et al,18 who demonstrated that when the DMT1 G185R mutant is stably expressed in cell lines it is abnormally glycosylated, less stable, and less active in metal transport. If present in detectable amounts in cells, the protein product of the exon 12–deleted mRNA does not appear to have a deleterious effect on iron absorption or utilization, as the patient's sister, who is a heterozygote and has a significant amount of the exon-deleted message, has no evidence of abnormal iron metabolism.
The phenotype of the patient in our study differs from that described in the mk mouse and the Belgrade rat; however, the rodent mutation results in an amino acid substitution whereas the effect of the human mutation is exon 12 skipping. While both rodent models of DMT1 mutation exhibit severe anemia as well as an impairment in iron absorption, our patient has severe anemia, but is iron overloaded. Thus in the patient there seems to be a discrepancy between iron absorption in the duodenum as compared with iron utilization in the erythroid cells. One plausible explanation for this observation is that the patient's DMT1 mRNA levels are greatly increased in the duodenum as compared with controls, such that the small percentage of full-length transcript is sufficient to permit adequate or even increased iron absorption. This does not seem to be the case, as total DMT1 transcript measured by real-time PCR was in the same range as the control duodenal samples. However, it should be noted that the levels of DMT1 mRNA varied approximately 15-fold within the control duodenum samples, with the patient mRNA level approximately 2-fold higher than the lowest control duodenal sample, but only one-seventh that of the highest sample (which was obtained from a patient with iron deficiency, a condition known to increase DMT1 message and protein levels). Western blots of the patient's duodenal DMT1 suggest that there may be additional control of expression at the translational or posttranslational level. Hepcidin level was measured in the urine of our patient and found to be low (4 ng/mg creatinine, normal range 10 ng/mg-200 ng/mg creatinine), consistent with her anemia. This may help to explain increased intestinal iron absorption in this patient; however, until more is known about how hepcidin regulates expression of DMT1 and other proteins involved in intestinal iron absorption, these data are difficult to interpret. Alternatively, there may be mechanisms of iron absorption in the duodenum that bypass DMT1. One such mechanism is the absorption of heme iron, a process that has not yet been well characterized. Heme iron is poorly absorbed by rats and mice, but in meat-eating humans it is estimated that about two-thirds of the absorbed iron is from heme.19 Thus in humans, a mutation in DMT1 protein may affect primarily iron utilization and not absorption, resulting in a different phenotype than that seen in the mk mouse and Belgrade rat. A third alternative is that iron absorption at the basolateral surface of the enterocyte is increased, that is, ferroportin or hephaestin. Unfortunately, the small samples of duodenum obtained at biopsy did not permit examination of this possibility.
Exon skipping and alternative splicing have been identified as primary mechanisms by which a relatively small number of genes are able to produce a complex proteome. In fact, it has been estimated that as many as 50% of all point mutations disrupt normal splicing.20 Exon skipping and alternative splicing appear to be a common theme in processing of DMT1 mRNA, as alternative exons at both the 5′ and 3′ ends of the molecule have been identified as well as skipping of exons 10 and 12.6,8 The function of exon skipping in the processing of the DMT1 message is unknown, as skipping of either exon 10 or 12 would be predicted to produce a protein that could not insert properly into the membrane.6 Exon 12–skipping appears to result from alteration of the consensus sequence for binding of the U1snRNP at the exon 12/intron 12 boundary; however, search of an exon-splicing enhancer database suggests that alterations in splicing enhancer regions may also be involved.21 Exon 12–skipping appears to be tissue specific, as only the full-length message is produced in normal duodenum, EBV-transformed lymphocytes, and CFU-GMs, but a small amount of exon 12–skipping is readily detectable in normal reticulocytes and to a lesser degree in granulocytes, monocytes, and platelets. It is possible that skipping of this exon is a developmental remnant and serves no useful purpose in the adult organism. Nevertheless, it may provide additional insight into the mechanisms of exon splicing by helping to identify exon-splicing enhancer and/or suppressor elements and the involved proteins and small RNA molecules.20,21
Prepublished online as Blood First Edition Paper, September 30, 2004; DOI 10.1182/blood-2004-07-2966.
Supported by National Institutes of Health training grant T32 DK60445 (M.P.M.), a VA Merit Review Grant and R01HL50077-11 (J.T.P.), and the Czech Republic Ministry of Health grant NM/6739-3 and MSM151100001 (D.P., M.P., K.I., V.D.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Jiri Ehrmann (Department of Pathology, Palacky University, Olomouc) for photomicrographs of the liver. We also thank Tomas Ganz and Elizabeta Nemeth for measurement of urinary hepcidin in our patient.