Abstract
Diamond-Blackfan anemia (DBA), a congenital erythroblastopenia, is a model disease for the study of erythroid differentiation but is poorly understood. RPS19 is the only gene yet to have been associated with DBA, but its relevance to erythroid differentiation is unclear. The molecular basis for the stimulation of erythropoiesis by glucocorticoids in patients with DBA has not been identified. We demonstrate that targeted degradation of the RPS19 transcript, through retroviral expression of short hairpin RNAs (shRNAs), blocks the proliferation and differentiation of erythroid progenitor cells in cultured human CD34+ cells. Treatment of RPS19-deficient cells with dexamethasone restores erythroid differentiation to normal levels. We investigated the molecular basis of pharmacologic therapies for DBA using oligonucleotide microarrays to survey gene expression in CD34+ cells treated with combinations of dexamethasone, erythropoietin, stem cell factor, and interleukin-3. Dexamethasone did not alter expression of RPS19 but activated a genetic program that includes a set of key hematopoietic regulatory genes. Genes specific to erythroid progenitor cells were up-regulated by dexamethasone, while genes specific to nonerythroid lineages were down-regulated. Deficiency of RPS19 therefore blocks proliferation of immature erythroid progenitor cells, and dexamethasone activates proliferation of the same cell population through mechanisms independent of RPS19. (Blood. 2005;105:4620-4626)
Introduction
Diamond-Blackfan anemia (DBA) is a rare congenital anemia characterized by erythroid hypoplasia, growth retardation, and other congenital anomalies.1-3 The molecular pathophysiology of DBA, leading to an almost complete loss of erythroid precursor cells in the bone marrow, is not understood. Insights into the basis of DBA promise not only to direct the development of novel therapeutic strategies for DBA, but also to inform the study of stem cell commitment and erythroid differentiation.
The only gene that has thus far been associated with DBA encodes ribosomal protein S19 (RPS19),4 while mutations in at least 2 other genes are likely to cause the DBA phenotype.5 The mechanisms whereby mutations in RPS19, a component of the small ribosomal complex, inhibit erythropoiesis are unknown. Mutations occur in approximately 25% of patients with DBA. Large deletions of one allele6-8 and certain frameshift and nonsense mutations lead to haploinsufficiency9 of RPS19. Taken together with reports that other missense mutations are scattered throughout the RPS19 gene,6,10,11 these data favor the notion that allelic insufficiency can cause DBA. Mice with homozygous inactivation of RPS19 are not viable, and although mice with inactivation of one allele were reported to have a normal hematocrit, RPS19 mRNA appears to be normal.12 It remains unclear whether partial loss of RPS19 function, through allelic insufficiency, is sufficient to cause the DBA phenotype.
Glucocorticoids are the primary pharmacologic therapy for patients with DBA, increasing red blood cell production in more than 50% of patients and increasing the proliferation of erythroid progenitor cells in vitro.13,14 However, the basis for the activity of dexamethasone in erythropoiesis has not been elucidated.
We investigated the effects of decreased RPS19 levels on erythropoiesis using RNA interference (RNAi). We targeted RPS19 mRNA transcripts for degradation in normal human hematopoietic progenitor cells and assayed the resulting changes in erythroid differentiation in vitro. We used this model system to study the activity of glucocorticoids in DBA. Furthermore, we used oligonucleotide microarrays to identify molecular targets of dexamethasone in primary human hematopoietic progenitor cells.
Materials and methods
Retroviral production and infection
Sequences targeted by short hairpin RNAs (shRNAs) are listed in Figure 1. shRNAs were cloned into pRETRO-SUPER, a self-inactivating, murine stem cell virus (MSCV)-derived, puromycin-expressing retroviral vector.15 Vesicular stomatitis virus-G-pseudotyped retrovirus was produced by cotransfection of retroviral vectors with packaging plasmids into 293 EBNA cells (Invitrogen, Frederick, MD). Primary CD34+ cells and HEL cells were plated one day prior to retroviral infections. Retroviral infections were performed in the presence of 8 μg/mL polybrene. At 24 hours after infection, retrovirally infected cells were selected using 1 μg/mL puromycin (Sigma, St Louis, MO).
The luciferase shRNA, cloned into an identical vector as the test shRNAs, was used as a control because it does not target any human genes but decreases expression of an exogenous luciferase gene and therefore engages the Dicer and RNA-induced silencing complex (RISC) complexes.16
Culture of primary and transformed cells
Human bone marrow CD34+ cells used in RNAi experiments were obtained from Cambrex (Poietics; Cambrex, East Rutherford, NJ). Erythroid differentiation was induced in 2 steps in liquid culture. For the first 7 days, cells were cultured in Serum-Free Expansion Medium (SFEM; Stem Cell Technologies, Vancouver, BC) supplemented with penicillin/streptomycin, glutamine, 25 ng/mL stem cell factor (SCF), 10 ng/mL interleukin-3 (IL-3), 10 ng/mL interleukin-6 (IL-6), and 0.5 IU/mL erythropoietin (Epo). After 7 days, cells were cultured in the same medium supplemented with 3 IU/mL Epo. Dexamethasone (Sigma) was added following retroviral infection at a concentration of 1 μM. Myeloid differentiation was induced in SFEM containing 25 ng/mL SCF, 10 ng/mL IL-3, 40 ng/mL Flt-3 ligand, and 15 ng/mL granulocyte colony-stimulating factor (G-CSF). SCF, IL-3, and IL-6 were obtained from Stem Cell Technologies; Flt-3 ligand was obtained from R&D Systems (Minneapolis, MN); and Epo (epoetin) and G-CSF (filgrastim) were obtained from Amgen (Thousand Oaks, CA).
For oligonucleotide microarray experiments, normal adult bone marrow was obtained from volunteer donors under a Dana-Farber Cancer Institute (DFCI) protocol. Mononuclear cells were collected from the interface after Ficoll Hypaque separation. Lineage-negative cells were collected after separation on a magnetic bead column (Dynal, Biotech, Brown Deer, WI), and CD34+ cells were purified by fluorescence-activated cell sorting. One sample was processed immediately for RNA preparation, and the remaining aliquots were cultured in the presence of either Epo (2 IU/mL) alone, Epo + IL-3 (20 ng/mL), or Epo + SCF (50 ng/mL), for 18 or 48 hours. Each growth factor combination was incubated in the presence or absence of 10-8 M dexamethasone. We chose this low concentration of dexamethasone for the gene expression studies because some DBA patients are extremely sensitive to very low doses of prednisone and because this concentration has been shown to be very effective in synergy with SCF in erythroid expansion studies.17,18 Cells cultured for 10 or 15 days were cultured in the 2-step erythroid differentiation conditions described in the previous paragraph.
K562, HEL, and TF-1 cells were obtained from American Tissue Culture Collection (ATCC, Manassas, VA). K562 and HEL cells were cultured in RPMI supplemented with 10% fetal bovine serum, penicillin/streptomycin, and glutamine. TF-1 cells were cultured in identical medium with the addition of 1 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF, filgrastim; Amgen).
Real-time RT-PCR
RNA was purified using Trizol (Life Technologies, Bethesda, MD). cDNA was produced with the Superscript II reverse transcriptase (Invitrogen) using oligo(dT) primers. Primers and probes for real time reverse transcriptase-polymerase chain reaction (RT-PCR) were obtained from Applied Biosystems Assays: gene expression Assay ID Hs00357218_g1 for RPS19, Hs00231112_m1 for GATA1, 401846 for β-actin, and Hs00735285 for RPS14 (Applied Biosystems, Foster City, CA). To calculate shRNA efficacy, RPS19 mRNA levels in cells expressing an RPS19 shRNA were compared with RPS19 levels in cells expressing a control shRNA. β-Actin was used as an endogenous control for infections of HEL cells. CD34+ cells infected with RPS19 shRNAs had a severe growth defect and expressed β-actin at very low levels, so the RPS14 gene was used as the endogenous control. Each real-time PCR reaction was performed in triplicate. Each knockdown experiment was performed at least twice, and the results of representative experiments are reported.
Western blot analysis
Cells were washed with phosphate-buffered saline and resuspended in lysis buffer containing 10 mM Tris (tris(hydroxymethyl)aminomethane), 100 mM NaCl, 1 mM EDTA (ethylenediaminetetraacetic acid), 1% triton-X, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS). Blots were blocked with 5% nonfat dry milk in Tris-buffered saline plus 0.1% Tween 20 and probed with a rabbit antibody against RPS199 at a concentration of 0.50 μg/mL and tubulin (Ab-4; Lab Vision, Fremont, CA) at a concentration of 0.75 μg/mL. Binding of horseradish peroxidase-linked secondary antibodies was visualized using the enhanced chemiluminescence (ECL) protein detection system (Amersham, Arlington Heights, IL).
Flow cytometry
Cells were harvested for flow cytometry 4 days after retroviral infection, 3 days after selection with puromycin. Approximately 1 to 5 × 105 cells were labeled with a phycoerythrin-conjugated antibody to glycophorin-A (Gly-A, CD235a, Clone GA-R2; BD-Pharmingen, San Diego, CA). Flow cytometry was performed using a FACScan flow cytometer from Becton Dickinson (San Jose, CA).
Methylcellulose colony assays
At 3 days after retroviral infection, and 2 days after selection with puromycin, cultured hematopoietic cells were plated in methylcellulose containing Epo, SCF, GM-CSF, and IL-3 (H4434; Stem Cell Technologies) plus 1 μg/mL puromycin in 35-mm plates. Colony formation was assessed after 14 days of culture at 37°C in a humidified atmosphere with 5% CO2. Evaluation of the colony assays was performed by an individual blinded to the experimental conditions. Since the methylcellulose cultures were plated after CD34 sorting and 5 days in liquid culture, we found that the erythroid colonies were virtually all mature erythroid burst-forming units (BFU-Es, small single or multicentric hemoglobinized colonies) at day 14. Similarly, granulocyte-macrophage colony-forming units (CFU-GMs) were smaller than obtained from cultures plated on day 0, and therefore granulocyte, monocyte, and granulocyte-monocyte colonies of more than 10 cells were scored as positive. Cells processed in parallel that were not retrovirally infected, but were exposed to puromycin in liquid and methylcellulose culture, did not produce any colonies.
DNA microarrays
RNA was prepared using Trizol (Life Technologies) in the presence of 5 μg linear polyacrylamide (GenElute LPA; Sigma) and 100 ng yeast tRNA (Roche Applied Sciences, Indianapolis, IN). RNA was linearly amplified by 2 rounds of in vitro transcription (IVT) by the method of Hunter and colleagues, modified to maintain representation of mRNA levels from small starting quantities of RNA and avoid template-independent amplification (Baugh et al19 ).
Briefly, first-round amplification was carried out with oligo(dT)-T7 primers and Superscript II (Invitrogen) in the presence of the single-stranded nucleic acid binding protein T4gp32 (USB) for one hour at 4°C. After second-strand synthesis in the presence of DNA polymerase, Escherichia coli RNase H, and E coli DNA ligase (Invitrogen, Cleveland, OH), the DNA was polished with T4 DNA polymerase and purified by phenol/chloroform extraction followed by chromatography on a BioGel P-6 MicroSpin Column (BioRad, Hercules, CA). After precipitation, IVT was performed (Ribomax kit; Promega), and the IVT products were purified using a Qiagen RNeasy Mini Column (Qiagen, Valencia, CA). For second-round amplification, random hexamer primers were added to the cRNA and RT was carried out at 37°C (20 minutes), 42°C (20 minutes), 50°C (10 minutes), and 55°C (10 minutes), and heat inactivated at 65°C (15 minutes). After RNAse H addition and annealing of oligo(dT)-T7 primer, second strand was synthesized without E coli ligase. After processing as for the first round, biotin-labeled aRNA was prepared using the Enzo kit (Affymetrix, Santa Clara, CA). A detailed protocol is described in the Document S1 (available on the Blood website; see the Supplemental
Results
Efficacy of RPS19 shRNA retroviruses
Approximately 25% of patients with DBA have a mutation in one allele of the gene encoding RPS19, but the functional consequences of most mutations are unknown, and the role of RPS19 in erythroid differentiation is not clear. To test whether the level of RPS19 expression is critical for erythroid differentiation, we designed retroviruses expressing RPS19 shRNAs that target RPS19 mRNA transcripts for degradation.
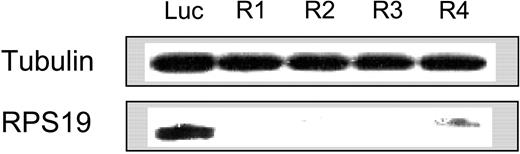
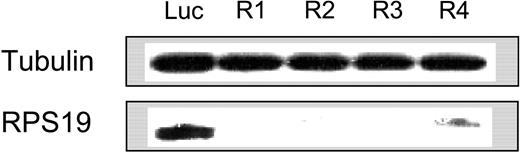
We generated 4 shRNA retroviruses that target different regions of the RPS19 gene (RPS19-R1 to RPS19-R4) and an shRNA targeting GATA-1, a transcription factor critical for erythroid differentiation. In order to test the efficacy of each shRNA in a homogeneous cell population that is not undergoing differentiation, we infected HEL cells, a erythroleukemia cell line, with the shRNA retroviruses. As shown in Table 1, each shRNA decreased expression of the RPS19 gene relative to the luciferase shRNA control by more than 50%. RPS19 shRNAs also decreased expression of the RPS19 protein as demonstrated by Western blot (Figure 1). The shRNAs decreased RPS19 expression in CD34+ cells as well, by 94%, 86%, and 84%, respectively, for RPS19-R2, RPS19-R3, and RPS19-R4.
RPS19 shRNAs block erythroid differentiation
The central phenotypic abnormality in patients with DBA is a deficiency of erythroid precursor cells in the bone marrow. We tested the effects of RPS19 shRNAs on hematopoietic progenitor and precursor cells using 2 in vitro assays of erythroid differentiation. First, we assayed expression of Gly-A, an erythroid-specific cell surface protein, by flow cytometry after induction of erythroid differentiation in liquid culture. Second, we analyzed the effects of RPS19 shRNAs on the formation of hematopoietic colonies after culture in methylcellulose.
The 2-phase induction of erythroid differentiation in liquid culture results in a synchronized differentiation of primary CD34+ cells with gradual up-regulation of Gly-A expression over the course of the 7-day experiment. As shown in Figure 2A, mean Gly-A expression did not vary significantly between cells infected with control shRNAs targeting the luciferase gene or cells infected by the empty vector retrovirus. The GATA1 shRNA, encoding a transcription factor that is critical for erythroid differentiation, decreased mean Gly-A expression to 53% plus or minus 6% of the control. All 4 RPS19 shRNAs decreased mean Gly-A expression, most powerfully by the RPS19-R1 shRNA, which decreased Gly-A to 57% plus or minus 13%. When cultured in the presence of cytokines promoting myeloid differentiation, the induction of CD11b was mildly decreased by expression of RPS19 shRNAs (Figure 2B). For each of the RPS19 shRNA retroviruses, the decrease in GlyA expression was statistically significant, with P less than .01 by a 2-tailed Student t test, while the decrease in CD11b was not statistically significant, with P greater than .10.
Expression of RPS19 shRNAs severely decreased the formation of all hematopoietic colonies after culture in methylcellulose (Figure 2C-D). The suppression of both BFU-E colony formation was statistically significant with P less than .001 for each RPS19 shRNA; the statistical significance of the decrease in CFU-GM colony formation was less dramatic, with P equal to .01 for RPS19-R1, P equal to .02 for RPS19-R2, and P equal to .04 for RPS19-R4. The decrease in erythroid colony formation was approximately twice as great as the decrease in myeloid colony formation. In contrast to RPS19 shRNAs, GATA1 shRNAs decreased erythroid colony formation but did not decrease the number of CFU-GM colonies (Figure S1).
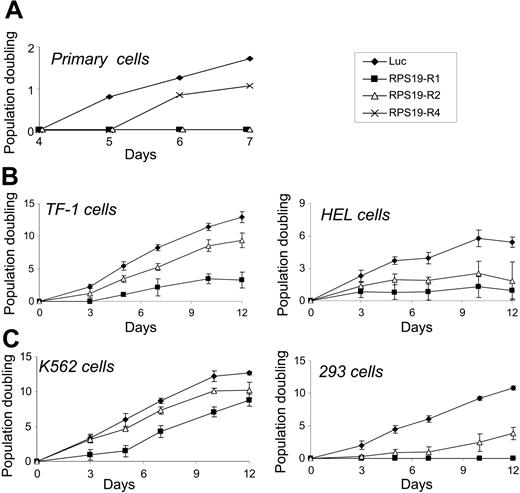
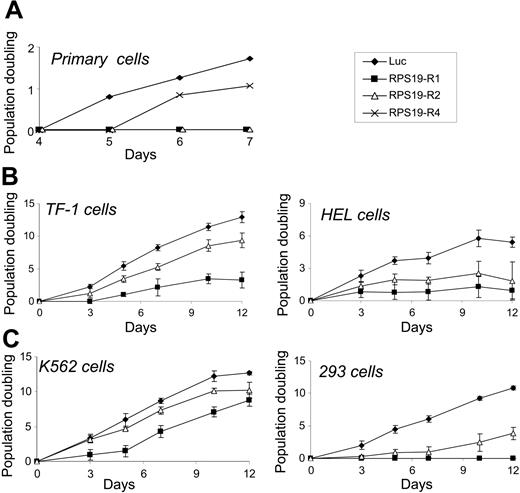
Differentiation and proliferation are intimately linked in erythropoiesis. The effects of RPS19 shRNAs on Gly-A expression and hematopoietic colony formation could be caused by impaired proliferation of erythroid progenitor cells. To assess proliferation independent of differentiation, we expressed RPS19 shRNAs in transformed cell lines. As shown in Figure 3, RPS19 shRNAs impaired the proliferation of CD34+ cells, 3 hematopoietic cell lines (HEL, K562, and TF-1), as well as 293 cells, an embryonic kidney cell line. The dramatic decrease in methylcellulose colony formation by RPS19-deficient cells may be due, at least in part, to a severe proliferative defect that prevents the formation of mature colonies.
Dexamethasone rescues erythroid differentiation in RPS19-deficient cells
Dexamethasone is the most effective pharmacologic therapy for patients with DBA. We investigated whether dexamethasone could correct the functional defect in erythroid differentiation caused by RPS19 shRNA expression in primary human hematopoietic cells.
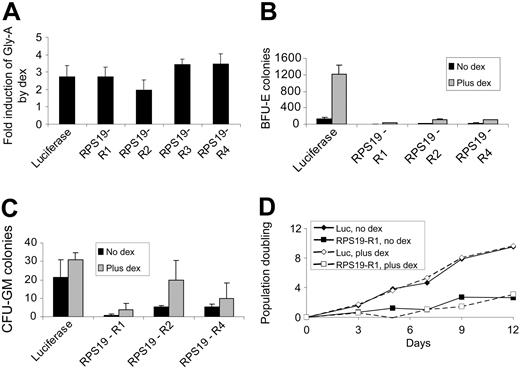
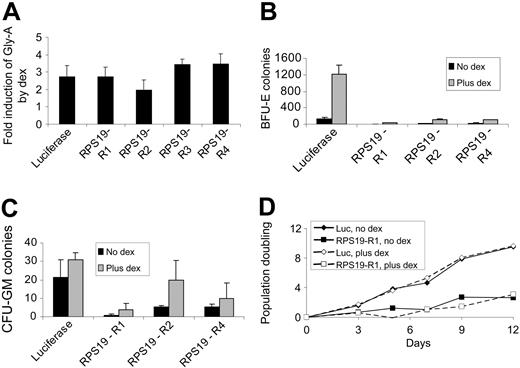
Dexamethasone was added to the liquid culture medium after infection of CD34+ cells with shRNA retroviruses. While mean Gly-A expression was decreased in cells expressing RPS19 shRNAs, treatment with dexamethasone increased mean Gly-A expression to a similar degree, 2.0- to 3.5-fold, in cells expressing the luciferase shRNA control and each of the RPS19 shRNAs (Figure 4A). The increase in Gly-A expression in cells treated with dexamethasone was sufficient to correct the decrease in Gly-A expression associated with each RPS19 shRNA. The effects of dexamethasone were independent of RPS19 expression, as Gly-A levels increased to a similar extent in control cells and in cells expressing each of the RPS19 shRNAs.
shRNA retroviruses decrease expression of RPS19. The effect of RPS19 shRNA retroviruses on RPS19 protein levels was evaluated by Western blot.
shRNA retroviruses decrease expression of RPS19. The effect of RPS19 shRNA retroviruses on RPS19 protein levels was evaluated by Western blot.
Similarly, in methylcellulose colony assays, dexamethasone increased erythroid colony formation both in cells infected with the control shRNA and in cells infected with each RPS19 shRNA (Figure 4B). Dexamethasone increased the formation of BFU-E colonies to a greater extent than CFU-GM colonies (Figure 4C). For cells expressing the luciferase, RPS19-R2, and RPS19-R4 shRNAs, the relative increase in BFU-E compared with CFU-GM colony formation was statistically significant (P < .05 by 2-tailed Student t test).
Erythroid differentiation is inhibited in human CD34+cells expressing RPS19 shRNAs. In panel A, mean glycophorin A (Gly-A) expression, assayed by flow cytometry, is shown as a percentage of the luciferase (luc) shRNA control (mean and SEM). In panel B, mean CD11b expression is displayed as a percentage of the control. Methylcellulose colony assays were performed with cells expressing control or RPS19 shRNAs. The number of BFU-E colonies is presented in panel C, and the number of CFU-GM colonies is presented in panel D (mean [SEM]). *P < .001 by 2-tailed Student t test; **P < .05.
Erythroid differentiation is inhibited in human CD34+cells expressing RPS19 shRNAs. In panel A, mean glycophorin A (Gly-A) expression, assayed by flow cytometry, is shown as a percentage of the luciferase (luc) shRNA control (mean and SEM). In panel B, mean CD11b expression is displayed as a percentage of the control. Methylcellulose colony assays were performed with cells expressing control or RPS19 shRNAs. The number of BFU-E colonies is presented in panel C, and the number of CFU-GM colonies is presented in panel D (mean [SEM]). *P < .001 by 2-tailed Student t test; **P < .05.
Dexamethasone is known to increase the proliferation of immature erythroid progenitor cells in vitro.17 To study the role of dexamethasone in RPS19 deficiency independent of the activity of glucocorticoids on primary erythroid progenitor cells, we examined the proliferation of HEL cells expressing RPS19 shRNAs. As shown in Figure 4D, dexamethasone did not influence the proliferation of cells expressing the luciferase control shRNA. RPS19 shRNAs decreased the proliferation rate of HEL cells, but dexamethasone did not increase the growth rate, indicating that RPS19 activity was not increased by dexamethasone in RPS19-deficient HEL cells.
Effect of DBA treatment modalities on gene expression
Having demonstrated that dexamethasone increases erythroid proliferation and differentiation in cells with normal and decreased levels of RPS19, we investigated whether dexamethasone acts by increasing expression of the RPS19 gene, either relative to or coordinately with the expression of genes encoding other ribosomal proteins. In addition, we analyzed the effects of 2 other established or potential DBA therapies: IL-3, which increases red blood cell production in 15% of patients,22-24 and SCF, which for many patients normalizes erythroid colony formation in vitro.25-27 Epo was included in all culture conditions because patients with DBA uniformly have elevated levels of Epo, and dexamethasone improves the expansion and differentiation of erythroid progenitor cells from DBA patients in the context of Epo stimulation.14
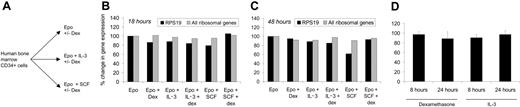
Primary cells expressing RPS19 shRNAs exhibited a severe proliferation defect. In order to focus on the effects of various therapies for DBA, independent of the effects of compromised proliferation, nonspecific effects of shRNA expression, and puromycin exposure, we analyzed the effects of the various treatments on normal human CD34+ cells. Cells were cultured with and without dexamethasone in the presence of Epo alone, Epo plus IL-3, or Epo plus SCF (Figure 5A). Gene expression was analyzed after 0, 18, and 48 hours of culture using Affymetrix U133A microarrays.
RPS19 shRNAs inhibit cellular proliferation. CD34+ cells cultured in erythroid differentiation medium (A), TF-1 cells (B), HEL cells (C), K562 cells (D), and 293 cells (E) were infected with control (luc) or RPS19 shRNAs and selected with puromycin. Cells were counted by hematocytometer and replated at a constant density.
RPS19 shRNAs inhibit cellular proliferation. CD34+ cells cultured in erythroid differentiation medium (A), TF-1 cells (B), HEL cells (C), K562 cells (D), and 293 cells (E) were infected with control (luc) or RPS19 shRNAs and selected with puromycin. Cells were counted by hematocytometer and replated at a constant density.
Dexamethasone promotes erythroid differentiation in cells expressing control and RPS19 shRNAs. (A) Human CD34+ cells expressing control (luc) or RPS19 shRNAs were grown in the presence or absence of dexamethasone (dex), and mean Gly-A expression was measured by flow cytometry. Fold induction by dexamethasone is shown as the mean and SEM. (B) Human CD34+ cells expressing control and RP19 shRNAs were grown with or without dexamethasone for 3 days and then plated on methylcellulose. The number of BFU-E colonies is shown as the mean and SEM. (C) CFU-GM colonies from the same cultures as panel B are shown as the mean and SEM. (D) HEL cells expressing the control (luc) or RPS19-R1 shRNA cultured in the presence (-) or absence (—) of dexamethasone were counted by hematocytometer and replated at a constant density.
Dexamethasone promotes erythroid differentiation in cells expressing control and RPS19 shRNAs. (A) Human CD34+ cells expressing control (luc) or RPS19 shRNAs were grown in the presence or absence of dexamethasone (dex), and mean Gly-A expression was measured by flow cytometry. Fold induction by dexamethasone is shown as the mean and SEM. (B) Human CD34+ cells expressing control and RP19 shRNAs were grown with or without dexamethasone for 3 days and then plated on methylcellulose. The number of BFU-E colonies is shown as the mean and SEM. (C) CFU-GM colonies from the same cultures as panel B are shown as the mean and SEM. (D) HEL cells expressing the control (luc) or RPS19-R1 shRNA cultured in the presence (-) or absence (—) of dexamethasone were counted by hematocytometer and replated at a constant density.
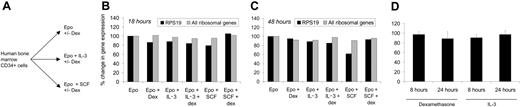
Expression of RPS19 was not influenced by dexamethasone, IL-3, or SCF (Figure 5B-C). The ribosomal protein genes were tightly coregulated across experimental conditions, and the average expression of these genes did not vary across experimental conditions. Using real-time RT-PCR, we confirmed that RPS19 expression is not altered in HEL cells treated with dexamethasone or IL-3 for 8 or 24 hours (Figure 5D). Finally, dexamethasone does not alter RPS19 expression in CD34+ cells expressing RPS19 shRNAs, increasing RPS19 expression by only 4% and 11% in cells expressing RPS19-R1 and RPS19-R2 shRNAs, respectively, as assessed by real-time RT-PCR.
DBA therapies do not regulate the expression of RPS19 or ribosomal genes. As illustrated in panel A, CD34+ cells were treated with Epo plus combinations of dexamethasone (dex), IL-3, and SCF, and gene expression was analyzed using oligonucleotide microarrays. In panels B and C, RPS19 expression and the mean expression of all ribosomal genes in CD34+ cells were assessed by microarrays. Gene expression for each treatment condition and time point is shown relative to gene expression in cells exposed only to Epo. In panel D, RPS19 expression was evaluated by real-time RT-PCR in HEL cells treated with dexamethasone or IL-3. RPS19 expression is depicted relative to untreated cells (mean and SEM).
DBA therapies do not regulate the expression of RPS19 or ribosomal genes. As illustrated in panel A, CD34+ cells were treated with Epo plus combinations of dexamethasone (dex), IL-3, and SCF, and gene expression was analyzed using oligonucleotide microarrays. In panels B and C, RPS19 expression and the mean expression of all ribosomal genes in CD34+ cells were assessed by microarrays. Gene expression for each treatment condition and time point is shown relative to gene expression in cells exposed only to Epo. In panel D, RPS19 expression was evaluated by real-time RT-PCR in HEL cells treated with dexamethasone or IL-3. RPS19 expression is depicted relative to untreated cells (mean and SEM).
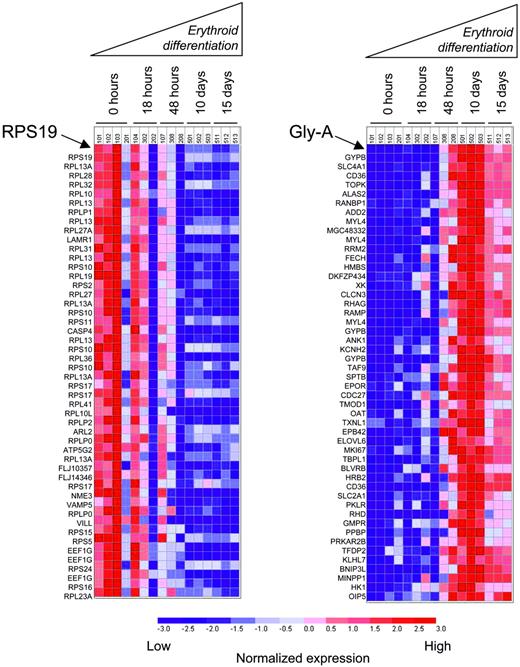
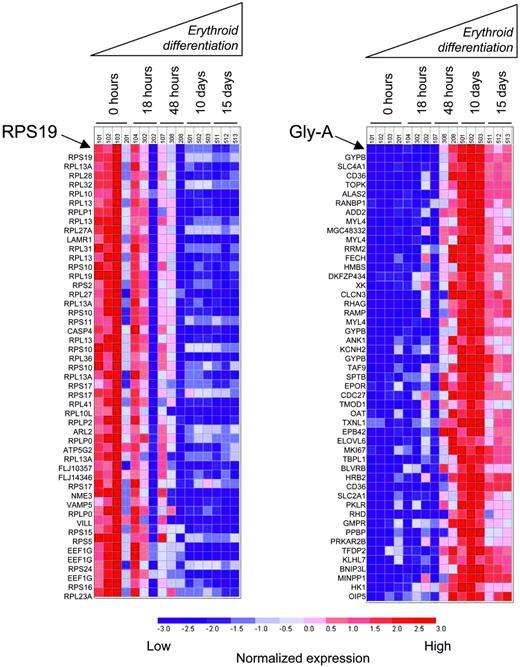
These experiments indicate that dexamethasone does not increase RPS19 expression. In fact, consistent with previous studies in murine erythroblast differentiation,28 RPS19 expression decreased in human hematopoietic progenitor cells during the course of erythroid differentiation in vitro (Figure 6). Furthermore, the expression of RPS19 was tightly coordinated with other ribosomal proteins. Using the Pearson correlation to rank the 22 284 probe sets on the U133A microarrays in order of the similarity in their expression profile across samples, the nearest neighbors of RPS19 were comprised almost entirely of ribosomal proteins and other proteins involved in translation. At a transcriptional level, RPS19 is not erythroid specific in its overall expression or in its expression relative to other genes encoding ribosomal proteins. Tight coregulation with other ribosomal proteins illustrates the primary role of RPS19 as a ribosomal protein and the need for precise stoichiometry in the assembly of ribosomes.
In contrast, the nearest neighbors of Gly-A are coordinately up-regulated during erythroid differentiation in the same samples (Figure 6). The nearest neighbors of Gly-A are genes encoding erythroid-specific proteins, including blood group antigens, components of the erythrocyte membrane skeleton, and enzymes involved in heme biosynthesis. The nearest neighbors of Gly-A do not include any ribosomal protein genes, and the nearest neighbors of RPS19 do not include any known erythroid-specific genes.
Taken together, these experiments indicate that the enhancement of erythropoiesis by glucocorticoids is independent of RPS19. While dexamethasone rescues erythroid differentiation in RPS19-deficient cells, the induction of erythropoiesis is independent of RPS19 levels. Dexamethasone, IL-3, and SCF do not alter the expression of RPS19 in hematopoietic progenitor cells. Moreover, while dexamethasone increases erythroid differentiation, RPS19 expression declines during erythroid differentiation.
Molecular targets of dexamethasone
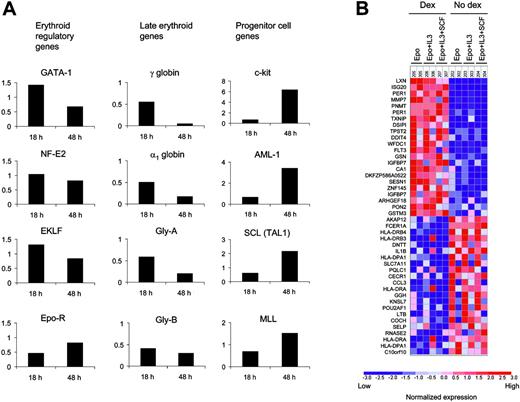
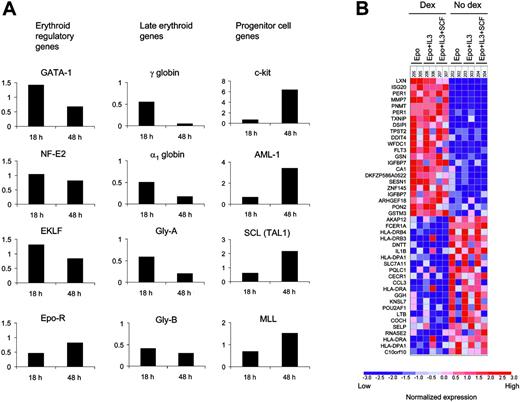
Erythropoiesis has been studied extensively, and key regulatory genes have been characterized.29 Dexamethasone did not increase expression of key erythroid regulators (Figure 7A) including GATA-1, nuclear factor-erythroid 2 (NF-E2), erythroid Kruppel-like factor (EKLF), and the erythropoietin receptor (EPO-R). Genes activated in terminal erythroid differentiation, including globin genes, were down-regulated in cells treated with dexamethasone for 18 or 48 hours. In contrast, genes expressed in hematopoietic progenitor cells, including c-kit, AML-1, SCL, and MLL, were up-regulated by dexamethasone. These changes were more pronounced at 48 hours, indicating that these changes may not be direct targets of dexamethasone.
RPS19 is down-regulated during erythroid differentiation. Rows correspond to the expression of individual genes. The 50 nearest neighbors of RPS19 (left) and Gly-A (right) are ranked by Pearson correlation. Columns represent independent microarray experiments of primary CD34+ cells undergoing erythroid differentiation in vitro stimulated by Epo and SCF. The dark red color indicates the highest gene expression, while the deep blue color reflects lowest gene expression. The nearest neighbors of RPS19 and Gly-A are listed in Tables S3 and S4, respectively.
RPS19 is down-regulated during erythroid differentiation. Rows correspond to the expression of individual genes. The 50 nearest neighbors of RPS19 (left) and Gly-A (right) are ranked by Pearson correlation. Columns represent independent microarray experiments of primary CD34+ cells undergoing erythroid differentiation in vitro stimulated by Epo and SCF. The dark red color indicates the highest gene expression, while the deep blue color reflects lowest gene expression. The nearest neighbors of RPS19 and Gly-A are listed in Tables S3 and S4, respectively.
In order to identify direct targets of dexamethasone, we examined global patterns of gene expression at the 18-hour time point. Dexamethasone engages the glucocorticoid receptor, a transcription factor, and would therefore be expected to activate a set of genes independent of the signaling cascades induced by different cytokines. The expression of 22 genes increased significantly in cells exposed to dexamethasone (P = .05 by permutation testing) across cytokine conditions (Figure 7B).
Targets of dexamethasone in CD34+cells were identified by oligonucleotide microarrays. (A) The effect of dexamethasone on the expression of key hematopoietic genes was analyzed in cells grown in the presence of Epo and SCF. The fold change in gene expression in response to dexamethasone at 18 and 48 hours of culture is shown for each gene. (B) Rows correspond to the genes most significantly up-regulated and down-regulated by dexamethasone across cytokine conditions. Each column is a different microarray experiment of CD34+ cells grown for 18 hours in the presence of the indicated cytokines with or without dexamethasone. Table S5 contains a list of genes induced by dexamethasone.
Targets of dexamethasone in CD34+cells were identified by oligonucleotide microarrays. (A) The effect of dexamethasone on the expression of key hematopoietic genes was analyzed in cells grown in the presence of Epo and SCF. The fold change in gene expression in response to dexamethasone at 18 and 48 hours of culture is shown for each gene. (B) Rows correspond to the genes most significantly up-regulated and down-regulated by dexamethasone across cytokine conditions. Each column is a different microarray experiment of CD34+ cells grown for 18 hours in the presence of the indicated cytokines with or without dexamethasone. Table S5 contains a list of genes induced by dexamethasone.
Many of the genes induced by dexamethasone are known to be expressed highly in immature hematopoietic progenitor cells or early erythroid precursors. For example, FLT3 is expressed highly in hematopoietic stem cells and itself promotes the proliferation of hematopoietic stem cells.30 Promyelocytic leukemia zinc finger (PLZF, ZNF145) is a transcriptional repressor that is expressed at high levels in hematopoietic progenitor cells, blocks hematopoietic differentiation by silencing target genes, and forms part of the PLZF/RARA fusion gene in some patients with acute promyelocytic leukemia.31 Carbonic anhydrase 1 (CA1)32 and gelsolin (GSN)33 are both known to be expressed at high levels in erythroid progenitor cells.
The genes most powerfully down-regulated by dexamethasone include some of the most specific markers of the lymphocyte, megakaryocyte, granulocyte, macrophage, and eosinophil lineages. Expression of 2 genes critical for lymphocyte development, OCT binding factor 1 (POU2AF1) and terminal deoxynucleotidyltransferase (DNTT), are decreased by dexamethasone. Expression of P-selectin (SELP), a component of platelet granules and a marker of megakaryocytes; eosinophil cationic protein (ECP, RNASE2), a component of eosinophil granules and a marker of eosinophils; and the Fc-component of immunoglobulin E (IgE, FCER1A), a marker of mast cells, are all decreased by dexamethasone. Major histocompatibility antigen (MHC) class II genes were down-regulated. Dexamethasone decreases expression of 3 cytokine genes, interleukin-1 beta (IL-1B), lymphotoxin-beta (LTB), and macrophage inflammatory protein-1-alpha (MIP-1α, CC chemokine ligand 3 [CCL3]), all of which are active in myeloid lineage cells.
Discussion
Mutations in the RPS19 gene have been identified in patients with DBA, but the role of these mutations in the pathophysiology of DBA has not been determined. We demonstrate, using retrovirally expressed shRNAs in cultured human CD34+ cells, that decreased levels of RPS19 are sufficient to cause a severe defect in the differentiation and proliferation of erythroid progenitor cells.
Glucocorticoids are an effective therapy for patients with DBA, but the mechanisms of action for dexamethasone and other DBA therapies are unknown. Dexamethasone increased erythroid differentiation in RPS19-deficient cells in vitro, but the effects of dexamethasone were independent of RPS19 levels. Furthermore, dexamethasone did not alter expression of the RPS19 gene or the genes encoding other ribosomal proteins.
We identified a set of genes that may be key mediators of the effects of dexamethasone. FLT3 is a tyrosine kinase that promotes the proliferation of hematopoietic stem cells,30 PLZF is a transcriptional repressor that blocks hematopoietic differentiation,31 and Mip-1α is a cytokine that blocks proliferation of hematopoietic stem cells.34 Induction of PLZF expression by dexamethasone in smooth muscle cells has been reported previously.35 By increasing expression of FLT3 and PLZF and decreasing expression of MIP-1A, dexamethasone may activate proliferation of hematopoietic progenitor cells and delay terminal differentiation. This is entirely consistent with reported in vitro effects of dexamethasone on increased erythroid proliferation and maturation delay of CD34+ cells.18 Furthermore, dexamethasone decreased expression of genes specific to nonerythroid hematopoietic differentiation while increasing expression of genes found in immature erythroid cells. Identification of the critical pathways activated by dexamethasone may aid in the development of novel therapies with improved activity for the treatment of DBA without the manifold side effects of glucocorticoids.
Our experiments are consistent with a model of DBA in which RPS19 deficiency decreases the rate of proliferation or increases the rate of apoptosis of erythroid progenitor cells. Dexamethasone corrects the defect by increasing proliferation of erythroid progenitor cells, the cells that are inhibited in DBA. Our data showing that dexamethasone acts independently of RPS19 expression are consistent with the efficacy of dexamethasone in patients without RPS19 mutations and continued associated abnormalities found in patients in remission on steroids such as high erythrocyte adenosine deaminase.
DBA has been termed a pure red blood cell aplasia, yet RPS19 has no clear role in erythropoiesis. In our experiments, expression of the RPS19 gene was down-regulated during erythropoiesis in coordination with other ribosomal protein genes. Functionally, RPS19 shRNAs block erythroid differentiation most powerfully, but also decrease formation of CFU-GM colonies and restrict proliferation of nonerythroid cells. Independent experiments performed concurrently to this work also demonstrated that RPS19 siRNA expression decreased both erythroid and myeloid colony formation in primary human hematopoietic cells.36 Nevertheless, RPS19 may have erythroid-specific function. The shRNAs used in our study decrease RPS19 expression to lower levels than in patients with DBA and may have more general effects due to disruption of ribosomal activity. In vivo, feedback regulation of the myeloid lineage may be able to compensate for RPS19 deficiency.
The phenotype of DBA patients is not restricted to the erythroid lineage. Previous studies suggest a defect in stem cells or uncommitted myeloid progenitors.37-39 In fact, many patients with DBA show progenitor deficiencies in vitro that suggest a defect in a multipotent progenitor. An age-related decline in BFU-Es and CFU-GMs has been described38,39 as well as a reduction in the erythroid and myeloid clonogenic output of long-term culture initiating cells (LTC-ICs).37,40,41 Patients with DBA have nonerythroid defects including growth retardation and can develop neutropenia later in life.40,41
Research on DBA has been limited by the lack of an easily accessible cell culture model of the disease. Partial inactivation of the RPS19 gene by RNAi creates a model of the disease that can be used to elucidate the mechanisms of RPS19 deficiency further and to test novel therapies for DBA. Targeted inactivation of other genes by RNAi in human hematopoietic progenitor cells can be used to test the function of other DBA candidate genes and to model other disorders of hematopoietic differentiation caused by decreased expression of a critical gene.
Prepublished online as Blood First Edition Paper, March 8, 2005; DOI 10.1182/blood-2004-08-3313.
Supported by the Howard Hughes Medical Institute (T.R.G.), and by the National Institutes of Health and the Marie Arturi Foundation (C.A.S.).
T.R.G. and C.A.S. codirected this work.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 2. Erythroid differentiation is inhibited in human CD34+ cells expressing RPS19 shRNAs. In panel A, mean glycophorin A (Gly-A) expression, assayed by flow cytometry, is shown as a percentage of the luciferase (luc) shRNA control (mean and SEM). In panel B, mean CD11b expression is displayed as a percentage of the control. Methylcellulose colony assays were performed with cells expressing control or RPS19 shRNAs. The number of BFU-E colonies is presented in panel C, and the number of CFU-GM colonies is presented in panel D (mean [SEM]). *P < .001 by 2-tailed Student t test; **P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/12/10.1182_blood-2004-08-3313/5/m_zh80120579690002.jpeg?Expires=1765007647&Signature=32MtDHrtOOQtR2~7zBIsCTPmHPqmyO4Jy2fnkwLxrpdFrPuOETwcIch2-spiarb~S0S2lt167LjCcXY4WzpedSD5YteB7X8gk-FZqaTu6a9b5Q-xZG4Cv5RoLTF5VbrYp8fLcSPZh1NF0VpIdO8aSshwvEYW0qT9S5TkJ6748PP-nN2wxqp050Nx3qVz4UlaffuYxV~qqYiChfzKgXm9qW1ixG5RFGSpDOcL4WXsAdZTDG~cLcOBrLhaHPCaMUk6ERMBGnw4ocaejNaq1JElwKRjkU23EIIMuQDka4QmqE9clyutyi-X10Ww7pN8JM67Wqpc8TNb3SQRTw8QvzepeQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. Erythroid differentiation is inhibited in human CD34+ cells expressing RPS19 shRNAs. In panel A, mean glycophorin A (Gly-A) expression, assayed by flow cytometry, is shown as a percentage of the luciferase (luc) shRNA control (mean and SEM). In panel B, mean CD11b expression is displayed as a percentage of the control. Methylcellulose colony assays were performed with cells expressing control or RPS19 shRNAs. The number of BFU-E colonies is presented in panel C, and the number of CFU-GM colonies is presented in panel D (mean [SEM]). *P < .001 by 2-tailed Student t test; **P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/12/10.1182_blood-2004-08-3313/5/m_zh80120579690002.jpeg?Expires=1765007648&Signature=sww8tbzQnwHvj6KcF2xJDzSUR4mfbRQoB9jgVq~INUR63XP92ux~6dzjO0x8p3~GVxFcxPQaiqyLmcqaEMqj-xpYN8N2WzaQReQapVOmuDrl28dWGNixxS2hD3wWWn2gRLyPIcsUuobkmMeOeUw7kATGU8pdHLbmfwTbRdlTbxFC7HqFdeEyiNeAItuJOmCww91gtgiDy8VFaVltUzRFyWoM~ffbQQGX6MhUpZMQKhd1p7kndujCzWEs8V2tRErBEvrqoyeVfa4xvZXxYVwjowEXVbuodSmlEIH4jecinbZlaCnF-tsQN1xVk6thz2J3Rb7JU7H1vrPO7j~ZgRgsEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)