Abstract
VE-cadherin is an adhesion molecule localized at the adherens junctions of endothelial cells. It is crucial for the proper assembly of vascular structures during angiogenesis and maintaining vascular integrity. We have studied 3 monoclonal antibodies (mAbs) against murine VE-cadherin that inhibit angiogenesis and tumor growth. Two of these, BV13 and 10G4, also disrupted normal vessels, resulting in severe vascular leakage, whereas the third, E4G10, did not. The goal of the current report was to identify the epitope of E4G10 and distinguish it from those of the disruptive mAbs. We mapped the epitope of E4G10 to within the first 10 amino acids of mature VE-cadherin and demonstrated that conserved tryptophan residues in this sequence are required for VE-cadherin–mediated trans-adhesion. The disruptive mAbs target a different epitope within amino acids 45 to 56, which structural homology modeling suggests is not involved in trans-adhesion. From our studies, we hypothesize that E4G10 can only bind the neovasculature, where VE-cadherin has not yet engaged in trans-adhesion and its epitope is fully exposed. Thus, E4G10 can inhibit junction formation and angiogenesis but is unable to target normal vasculature because its epitope is masked. In contrast, BV13 and 10G4 bind an epitope that is accessible regardless of VE-cadherin interactions, leading to the disruption of adherens junctions. Our findings establish the immediate N-terminal region of VE-cadherin as a novel target for inhibiting angiogenesis.
Introduction
Cadherins are a large family of adhesion molecules involved in the formation of specific cell-cell contacts.1 In humans, more than 80 members of the cadherin superfamily have been identified and are classified into subfamilies according to the presence of conserved domains and sequence motifs.2 Cadherins are single-pass transmembrane glycoproteins defined by distinctive extracellular cadherin domains (ECDs) of about 110 amino acids. They mediate calcium-dependent homophilic interactions and are responsible for selective cell-cell recognition and adhesion. These processes play an important role during embryonic morphogenesis and maintenance of tissue architecture.2,3
Cadherins are subdivided according to specific sequence features; subfamilies include the type I (eg, N-, E-, and C-) and type II (eg, VE- and MN-) cadherins. Both type I and type II cadherins consist of 5 ECDs (ECD1-5) and are anchored to the actin cytoskeleton through their cytoplasmic tail.4 The determinants of adhesion and adhesive specificity among the type I classic cadherins, specifically the N-, E- and C-cadherins, have been extensively studied.5-10 In particular, the 3-dimensional structures of the N-terminal domains of N- and E-cadherins as well as the entire 5 ECDs of C-cadherin have been solved. These data indicate that the crucial adhesive determinants reside in the N-terminal ECD1 with the central feature being a conserved tryptophan residue (W2) that inserts into the hydrophobic core of the partner cadherin molecule presented by a juxtaposed cell.5-7,9 However, studies using different approaches have suggested that cadherin binding and adhesion may involve other ECDs as well,11-17 but the exact nature of these interactions is not fully understood.
The vascular endothelial-specific VE-cadherin mediates homophilic adhesion between neighboring endothelial cells and is categorized as a type II cadherin due to the presence of 2 conserved tryptophan residues (W2, W4) in the N-terminal sequence.18 Although little is known about the mechanism of type II cadherin-mediated adhesion, it is generally thought that the overall 3-dimensional structures and the homophilic interactions of the type I and type II cadherins are similar, despite the low sequence homology between VE-cadherin and the type I N-, E-, and C-cadherins (∼25%).
VE-cadherin is localized within specialized structures at cell-cell contacts, called adherens junctions, and is constitutively expressed throughout the entire vasculature.19-21 Accumulating evidence implicates VE-cadherin in various aspects of vascular biology including endothelial cell migration,19 survival,22 contact-induced growth inhibition,23 vascular integrity,24-26 and, most notably, endothelial-cell assembly into tubular structures.27 The importance of VE-cadherin in developmental angiogenesis has been demonstrated by the severe impairment of vascular assembly in VE-cadherin null embryos, leading to embryonic lethality by day E9.5.22 Because VE-cadherin plays such a crucial role in embryonic neovascularization, antibodies that block VE-cadherin assembly may be effective agents to inhibit tumor angiogenesis and thus tumor growth. Indeed, monoclonal antibodies (mAbs) developed against various extracellular regions of VE-cadherin block adherens junction formation in vitro,26,28 disrupt established vessels,26 and inhibit vascular assembly in vivo, thereby blocking angiogenesis and tumor growth.26,29 These studies demonstrated the importance of VE-cadherin in postnatal angiogenesis and its importance in the maintenance of vascular integrity.
We have previously reported on the distinct functional effects of 2 mAbs against murine VE-cadherin (mVE-cadherin), BV13 and E4G10.26,29 Although both mAbs inhibit angiogenesis and tumor growth, BV13 disrupts the adherens junctions between established vessels, most notably in the lung, causing severe hemorrhaging and mortality. In contrast, E4G10 inhibits VE-cadherin–mediated adhesion without increased vascular permeability or toxicity.29 Using fluorescently labeled mAbs, we demonstrated that E4G10 binds to only the tumor neovasculature, whereas BV13 binds to all vasculature.29 These results demonstrate that specific targeting of VE-cadherin on tumor vasculature is possible, despite ubiquitous VE-cadherin expression in normal vessels. It, therefore, became of interest to us to evaluate the therapeutic potential of targeting VE-cadherin.
Given the complexity of VE-cadherin–mediated adhesion, we hypothesized that certain epitopes of the VE-cadherin molecule may be exposed to the local environment only prior to the formation of trans-adhesion dimers at endothelial cell-cell contacts. Such temporally exposed epitopes might be accessible to mAbs, and mAbs directed against these epitopes may be able to prevent cell-cell adhesion and thus inhibit angiogenesis, whereas other epitopes would be accessible regardless of adhesion and thus susceptible to disruption. We therefore set out to identify the epitope of E4G10 and distinguish it from those of the disruptive mAbs, as well as demonstrate the importance of these epitopes in adherens junction formation.
Materials and methods
Cells and media
The mouse endothelial cell line H5V (from Dr A. Vecchi, Instituto Mario Negri, Milan, Italy) and 293 cells were cultured in Dulbecco modified Eagle medium (DMEM) with l-glutamine, (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (HyClone Laboratories, Logan UT), and 1% penicillin-streptomycin (Invitrogen). CHO and COS cells were cultured in RPMI medium with l-glutamine (Invitrogen) containing 10% fetal bovine serum, and 1% penicillin-streptomycin.
Antibody generation
Antibody BV13 was generated as described.25 The other mAbs were generated by immunizing Lewis rats with either an affinity-purified soluble fusion protein made by joining the VE-cadherin ECD1-5 with human IgG1 Fc (VE-cadherin-Fc), or a synthetic 15mer peptide derived from the N-terminal domain of mVE-cadherin, corresponding to amino acids 1-15. The 15mer peptide was KLH-coupled at its C-terminus via an added cysteine residue (Quality Controlled Biochemicals, Hopkinton, MA). Supernatants from several hundred hybridoma clones were screened for binding to VE-cadherin-Fc or the synthetic peptide immobilized on enzyme-linked immunosorbent assay (ELISA) plates.29 Antibodies were purified from conditioned medium by protein G-Sepharose chromatography and contained less than 1.25 EU/mL endotoxin, as assessed by the Limulus amebocyte lysate assay kit (BioWhittaker, Walkersville, MA).
Antibody characterization
To assess our effectiveness of mAbs in blocking adherens junction formation, an in vitro permeability assay28 was performed using H5V endothelial cell monolayers cultured on Costar transwells (24 wells, 0.4-μm pore; Corning Costar, Cambridge, MA). Adherens junctions were disrupted by chelating calcium in the medium with 5 mM EGTA (ethylene glycol tetraacetic acid) for 15 minutes at room temperature. The effect of the mAbs on junctional reformation was quantified by measuring the permeability of the endothelial cell monolayer using fluorescein isothiocyanate (FITC)–dextran (40S, final concentration 1 mg/mL; Sigma, St Louis, MO) as a fluorescence read-out. FITC-dextran was added to the upper chambers 2 hours after addition of calcium and the test mAb, and aliquots from the bottom chamber of the transwells were removed 1 hour later for determination of the FITC-dextran content by fluorometry (Sequoia-Turner, Model 450, Las Vegas, NV) using absorption/emission wavelengths set at 492/520 nm. A wide range of mAb concentrations (0.1-200 μg/mL) was tested in this assay and the median inhibitory concentration (IC50) value for each mAb was obtained.
To assess the ability of our mAbs to block formation of capillary vessels in vivo, a Matrigel plug angiogenesis assay was performed as previously described,28 using 1-mg doses of E4G10, 13E6, and 15F12, and 50-μg doses of BV13 and 10G4 administered intraperitoneally 3 times per week.
In vivo permeability studies were performed to assess the disruptive activity of the mAbs. Various doses of the mAbs, ranging from 25 to 3000 μg/mouse, were administered intraperitoneally and tested for their effects on vascular permeability in lung tissue using a modified Miles assay (Evans blue extravasation), as previously described.29
Antibody domain mapping
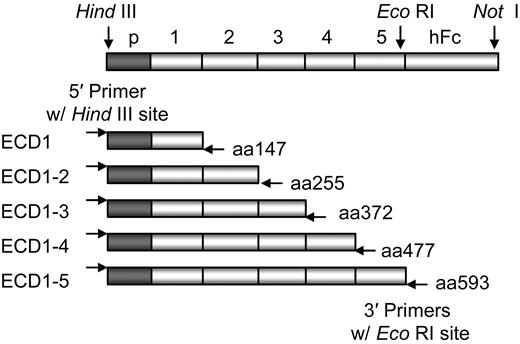
To identify the extracellular domain to which each mAb binds, soluble VE-cadherin-ECD-Fc fusion proteins were generated that differed by the number of ECDs contained in them (Figure 1). The chimeric plasmids were constructed using a polymerase chain reaction (PCR)–based strategy. PCR-generated mVE-cadherin ECD fragments were ligated into the expression vector pD10.105, which contained cDNA encoding the hinge and CH2 and CH3 regions of human IgG1. VE-cadherin ECD fusion proteins of 102, 210, 327, 432, and 548 amino acids were generated representing ECD1, ECD1-2, ECD1-3, ECD1-4, and ECD1-5 VE-cadherin fragments, respectively. All the constructs were confirmed by DNA sequencing and expression of the corresponding fusion proteins were examined by Western blot analysis using the supernatants collected from COS cells that were transiently transfected with each construct. Binding of the various mAbs to the fusion proteins in the COS supernatants was assessed by ELISA as previously described.29
mAb domain mapping. Diagram of the truncated soluble mVE-cadherin-Fc constructs ECD1, ECD1-2, ECD1-3, ECD1-4, and ECD1-5. The VE-cadherin prosequence (p) and the human Fc receptor (hFc) are labeled on the diagram along with the ECDs and fragment sizes.
mAb domain mapping. Diagram of the truncated soluble mVE-cadherin-Fc constructs ECD1, ECD1-2, ECD1-3, ECD1-4, and ECD1-5. The VE-cadherin prosequence (p) and the human Fc receptor (hFc) are labeled on the diagram along with the ECDs and fragment sizes.
VE-cadherin mutants
Plasmids encoding full-length murine and human (pVE3) VE-cadherin cDNA, cloned into the NheI-XbaI sites of pcDNA3.1(+)/Zeo, were used as templates to clone chimeric human-murine VE-cadherin expression plasmids. The chimeric expression plasmids were generated using a 2-step PCR strategy. To construct the human-murine VE-cadherin chimeras we obtained 2 human VE-cadherin cDNAs from the 5′ NheI site (including the prosequence) up through the nucleotides encoding amino acids 31 and 56, respectively, using PCR. We then cloned the corresponding mVE-cadherin cDNAs from a 3′ EcoRV site (created at the ECD1-2 junction) down through the nucleotides encoding amino acids 31 and 56, generating overlapping human and murine sequences. The overlapping human and mVE-cadherin DNA fragments were then pieced together through a second round of PCR amplification and cloned into the NheI-EcoRV sites of the mVE-cadherin ECD1 to generate human-mouse chimeric expression plasmids containing the human N-terminal sequence (amino acids 1-31 and 1-56, respectively) joined to the remaining portion of mouse VE-cadherin (human VE [hVE]–cadherin1-31/mVE-cadherin32-739 and hVE-cadherin1-56/mVE-cadherin57-739; Figure 3A). A similar approach was used to generate a chimeric VE-cadherin in which the amino acid sequence 1-56 of hVE-cadherin was replaced with the corresponding mouse sequence (mVE-cadherin1-56/hVE-cadherin57-737). Likewise, chimeras between murine N-cadherin (mN-cadherin) and mVE-cadherin were constructed (mN-cadherin1-10/mVE-cadherin11-739 and mN-cadherin1-15/mVE-cadherin16-739; Figure 2B).
Individual amino acid substitutions for the alanine scanning and human-murine VE-cadherin chimera substitution studies were generated by site-directed mutagenesis of the full-length mVE-cadherin expression vector pVE2 using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Each VE-cadherin substitution was confirmed by sequencing across the site of mutagenesis.
Identification of the epitope for E4G10. (A) Alanine scanning was performed in full-length mVE-cadherin cDNA expression plasmids, with substitutions at amino acid positions 1-15. Mock-transfected, as well as 293 cells expressing murine wild-type (mWT) and mutant mVE-cadherin expression plasmids were stained with E4G10 followed by goat anti–rat-PE secondary antibody to determine relative MFI by flow cytometry. The mAb-binding activity on the alanine-substituted VE-cadherin is measured relative to binding activity on mWT VE-cadherin and normalized for transfection efficiency (percent relative MFI, see “Materials and methods”). Bar graphs represent the average of 3 separate experiments with SDs. (B) Diagram of mWT and mN-cadherin1-10/mVE-cadherin11-739 (1-10 N/VE) and mN-cadherin1-15/mVE-cadherin16-739 (1-15 N/VE) chimeric molecules. VEC indicates VE-cadherin. (C) Results of E4G10 binding to the 1-10 N/VE and 1-15 N/VE chimeric molecules compared to mWT and mock transfected (n = 3).
Identification of the epitope for E4G10. (A) Alanine scanning was performed in full-length mVE-cadherin cDNA expression plasmids, with substitutions at amino acid positions 1-15. Mock-transfected, as well as 293 cells expressing murine wild-type (mWT) and mutant mVE-cadherin expression plasmids were stained with E4G10 followed by goat anti–rat-PE secondary antibody to determine relative MFI by flow cytometry. The mAb-binding activity on the alanine-substituted VE-cadherin is measured relative to binding activity on mWT VE-cadherin and normalized for transfection efficiency (percent relative MFI, see “Materials and methods”). Bar graphs represent the average of 3 separate experiments with SDs. (B) Diagram of mWT and mN-cadherin1-10/mVE-cadherin11-739 (1-10 N/VE) and mN-cadherin1-15/mVE-cadherin16-739 (1-15 N/VE) chimeric molecules. VEC indicates VE-cadherin. (C) Results of E4G10 binding to the 1-10 N/VE and 1-15 N/VE chimeric molecules compared to mWT and mock transfected (n = 3).
Determination of mAb binding to VE-cadherin mutants
Binding of the mAbs to wild-type and the various VE-cadherin mutants was assessed by flow cytometry to identify mAb epitopes. The plasmids encoding both wild-type and various VE-cadherin mutants were transiently transfected into 293 cells, in 6-well plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 48 hours after transfection the cells in each well were disassociated, split into tubes, incubated with each of the test mAbs, and next with a secondary goat anti–rat phycoerythrin (PE) antibody, and then analyzed by flow cytometry (Epics XL, Beckman Coulter, Hialeah, FL). The mean fluorescent intensity (MFI) of each mAb binding to the various VE-cadherin proteins was measured by the Beckman Coulter flow cytometry software (EXPO32ADC). The percent relative MFI was calculated by first dividing the MFI of the each mAb bound to the mutant VE-cadherins by the MFI of the same mAb bound to wild-type mVE-cadherin, to determine the relative MFI. Next, we normalized for transfection efficiency by dividing the relative MFI of each test mAb with the relative MFI of the ECD4-binding mAb 15F12 within each transient transfection (ie, each well).
Determination of junctional clustering of VE-cadherin mutants
Clustering of VE-cadherin to adherens junctions is an indicator of biologic activity of VE-cadherin in endothelial cells as well as in other cells expressing VE-cadherin.19 To assess junctional clustering, CHO cells were transfected with wild-type or mutant VE-cadherin expression plasmids using Lipofectamine 2000 and then selected with Zeocin (500 μg/mL, Invitrogen). VE-cadherin expression levels were determined by flow cytometry as described (see “Determination of mAb binding to VE-cadherin mutants”). Cells were grown as monolayers on chamber slides (Nalge Nunc International, Naperville, IL) and immunostained.28 Briefly, cells were fixed with 5% paraformaldehyde for 15 minutes, incubated with 10 μg/mL of the mAb 15F12 for 45 minutes, and then a secondary goat anti–rat-cyanine 3 (Cy3) antibody for 30 minutes. Cells were visualized using a Zeiss Axioskop microscope (40× objective), and the fluorescent images were captured with a Zeiss AxioCam using Zeiss AxioVision acquisition software (Carl Zeiss MicroImaging, Thornwood, NY).
Homology modeling
The amino acid sequences were taken from SWISS-PROT.30 First, sequence alignments of the extracellular cadherin domains (EC1-EC5) of C-cadherin and VE-cadherin were generated using CLUSTALX version 1.831 and TCOFFEE version 1.3732 with default parameters. Alternative user-adjusted alignments based on these were also made. Best efforts were made to align secondary structure elements of VE-cadherin as predicted by the PredictProtein server33 with the secondary structure elements in C-cadherin. C-cadherin (PDB: 1L3W), which served as the structural template, and each alignment were inputted into NEST34 to build a homology model of VE-cadherin. Next, WHATIF35 was used to verify the protonation states of histidine, aspartic acid, and glutamic acid residues as well as positions of oxygen and nitrogen atoms of asparagine, glutamine, and histidine side chains in each model. Hydrogen atoms were then built using CHARMM version 30a1.36 Following the addition of hydrogen atoms, each structure was energy minimized in vacuum first with a short run of steepest descent minimization (20 steps) and further with a conjugate gradient method of energy minimization until an energy gradient of 0.05 kcal/mol per A2 was reached. A harmonic constraint of 2 kcal/mol per A2 was applied to heavy atoms during the conjugate gradient minimization. Using Verify3D,37 we evaluated each model and selected the structure that had the best 3-dimensional profile. Lastly, for the best model, we identified amino acid residues that scored poorly according to the Verify3D profile and then refined each side chain using SCAP.38
Results
Characterization of mAbs against mVE-cadherin
In addition to the 2 previously characterized mAbs BV1325,26,29 and E4G10,29 we identified 5 mAbs that recognized mVE-cadherin (Table 1). All 7 mAbs bind soluble mVE-cadherin-Fc, as well as native mVE-cadherin molecules expressed on H5V cells, as determined by ELISA and flow cytometry, respectively (data not shown). The mAbs have similar binding affinities (5-15 nM; data not shown) as measured by binding assays using 125I-labeled mAbs and calculated by Scatchard analysis.
We used an in vitro permeability assay to mimic the formation of new adherens junctions and tested the ability of our mAbs to inhibit this process. For this purpose, the adherens junctions of endothelial cell monolayers grown on transwells were disrupted, and antibody-mediated inhibition of junction reformation was determined by measuring FITC-dextran passage through the endothelial monolayer after addition of the anti-VE-cadherin mAbs. A broad range of mAb concentrations was examined and IC50 values were obtained. BV13, 10G4, and E4G10 inhibited junction formation, whereas the mAbs 13E6, 8A7, 15F12, and 2B11 had no significant effect (Table 1).
We further tested the ability of the mAbs to inhibit the assembly of new capillary vessels in vivo using the Matrigel plug assay.28 Neovascularization was measured in the plugs of mice that received growth factor-supplemented Matrigel and administered a 50-μg (BV13, 10G4) or 1-mg (E4G10, 13E6, 8A7, 15F12) dose of mAb 3 times weekly. The mAbs E4G10, BV13, and 10G4 inhibited neovascularization in the plugs by 78%, 72%, and 89%, respectively (Table 1; P < .05). No significant inhibition of angiogenesis was detected with 13E6, 8A7, and 15F12 treatments (Table 1).
Both BV13 and E4G10 have been shown previously to bind mVE-cadherin and inhibit angiogenesis in vitro and in vivo.25,26,29 However, BV13 not only inhibits tumor angiogenesis but also disrupts normal vasculature, resulting in increased vascular permeability and associated toxicity.26,29 We therefore screened our new mAbs in vivo for their effect on vascular permeability and toxicity. Mice were administered a single dose of mAb ranging from 0.1 mg up to 10 mg, followed 20 hours later by an injection of Evans blue dye for assessment of dye extravasation into mouse organs. Treatment with 10G4 significantly increased dye extravasation in the lungs of mice (> 200%; Table 1) in a manner similar to BV13,29 and thus like BV13, it was designated as a “disruptive” mAb (Table 1). Pathohistologic examinations performed on the lungs of the mice treated with 10G4 demonstrated massive hemorrhage due to severe vascular leakage (data not shown). In contrast, as previously reported. the antiangiogenic mAb E4G10 had no effect on vascular permeability even at higher doses,29 and therefore was designated a “nondisruptive” mAb. The mAbs 13E6, 8A7, and 15F12, which had little or no activity in the in vitro permeability and Matrigel plug assays, had no activity in the permeability assay and were also designated nondisruptive (Table 1).
Epitope mapping
The ECDs to which the mAbs bind were initially mapped using a series of sequentially truncated soluble mVE-cadherin-Fc proteins consisting of ECD1, ECD1-2, ECD1-3, ECD1-4, and ECD1-5 (Figure 1). Expression of the truncated mVE-cadherin proteins in cell lysates was confirmed by Western blot analysis (data not shown). The ability of individual mAbs to bind the truncated mVE-cadherin proteins was examined by ELISA. The mAbs BV13, 10G4, and E4G10 bind mVE-cadherin ECD1 alone and are thus referred to as ECD1 mAbs. The mAbs 13E6, 8A7, 15F12, and 2B11 bind ECD2, ECD3, ECD4 and ECD5, respectively. Only those mAbs that bind ECD1 demonstrated in vitro and in vivo antiangiogenic activity (Table 1).
E4G10 binds ECD1 as expected, given that it was generated against the 15mer peptide corresponding to the N-terminal sequence of mVE-cadherin (amino acids 1-15). In an effort to identify individual amino acid residues involved in E4G10 binding, alanine scanning was performed within the first 15 amino acids, mutating individual amino acids to alanine in the context of the full-length mVE-cadherin sequence. The resulting mutated mVE-cadherins were expressed in 293 cells. E4G10-binding activity was measured on the panel of mutant mVE-cadherins by flow cytometry. Substitutions W4A, H8A, I9A, and D10A all resulted in a greater than 50% decrease in binding activity as compared to E4G10 binding to murine wild-type (mWT) VE-cadherin (Figure 2A). Lesser decreases in binding were observed for the N5A and M7A substitutions. These findings identify amino acids W4, N5, M7, H8, I9, and D10 as a potential E4G10-binding epitope.
We further demonstrated that the first 10 amino acids of mVE-cadherin are sufficient for E4G10 binding by measuring mAb-binding activity on chimeric cadherin molecules that incorporated murine N-cadherin (mN-cadherin) amino acids into full-length mVE-cadherin. Because E4G10 does not bind mN-cadherin we generated mN-cadherin1-10/mVE-cadherin11-739 and mN-cadherin1-15/mVE-cadherin16-739 chimeras, respectively (Figure 2B), and measured mAb-binding activity to them. We tested binding to both chimeras because E4G10 was generated from animals immunized with a peptide composed of amino acids 1-15, even though the results of our alanine scanning studies identified the epitope to be within the first 10 residues. We observed the complete loss of E4G10 binding to both chimeras (Figure 2C). These data confirm that the epitope of E4G10 is contained within the first 10 amino acids of mVE-cadherin.
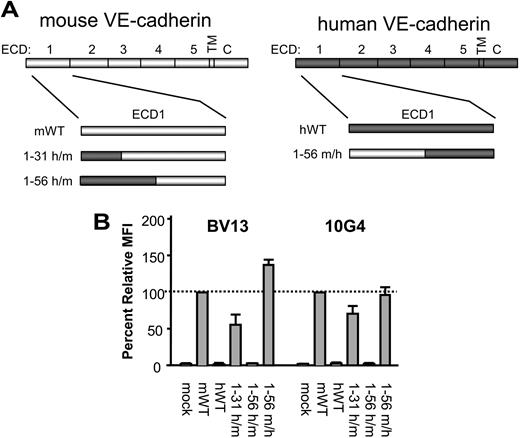
To map the epitopes of the disruptive mAbs, we took advantage of a previous observation that BV13 and 10G4 bind to mVE-cadherin but not hVE-cadherin. Because we have previously shown these mAbs to bind to the ECD1, we tested chimeric proteins in which increasingly larger regions of the murine ECD1 sequence were replaced with the corresponding human ECD1 sequence. We tested mAb-binding activity on chimeric VE-cadherin molecules that incorporated hVE-cadherin amino acids 1-31 and 1-56 into full-length mVE-cadherin, generating hVE-cadherin1-31/mVE-cadherin32-739 and hVE-cadherin1-56/mVE-cadherin57-739 chimeras, respectively (Figure 3A). BV13 and 10G4 binding to the hVE-cadherin1-31/mVE-cadherin32-739 chimera was reduced to 56% and 71%, respectively, and was almost completely abolished for both mAbs with the hVE-cadherin1-56/mVE-cadherin57-739 (Figure 3B). We then performed the reverse experiment, swapping in the first 56 amino acids of mVE-cadherin into hVE-cadherin to generate a mVE-cadherin1-56/hVE-cadherin57-737 chimera (Figure 3A), and looked for regained mAb-binding activity. Indeed, both BV13 and 10G4 bind the mVE-cadherin1-56/hVE-cadherin57-737 chimera at levels similar to that observed on wild-type mVE-cadherin (Figure 3B). Therefore, the epitopes for the disruptive mAbs appear to include residues within the first 56 amino acids of mVE-cadherin.
Because the disruptive mAbs BV13 and 10G4 bind the mVE-cadherin1-56/hVE-cadherin57-737 chimera but not the hVE-cadherin1-56/mVE-cadherin57-739 chimera protein, our findings indicate that some of the nonconserved residues within the N-terminal 1-56 amino acids constitute a part of the binding epitopes for these antibodies. A comparison of the first 56 amino acids of human and murine VE-cadherin reveals that there are 14 nonconserved amino acids between them (Figure 4A). By swapping 2 or 3 of these residues at a time from mouse to human, and measuring the loss of mAb-binding activity, we further attempted to localize the disruptive mAb epitopes. For this purpose, we generated 6 additional human-murine chimeras by replacing mVE-cadherin sequences 15-20, 27-31, 36-38, 41-42, 45-51, and 54-56 with those of hVE-cadherin (Figure 4A). In this manner, all nonconserved residues of the 1-56 amino acid sequence are represented in one of these chimeras. We observed the greatest loss of binding activity for both mAbs on chimeras that replaced mVE-cadherin residues 45-51 and 54-56 with those of hVE-cadherin (Figure 4B). Therefore, the disruptive mAbs appear to be targeting an epitope on mVE-cadherin that includes nonconserved residues within amino acids 45-56.
Identification of the disruptive mAb-binding domain. (A) Diagram illustrates the hVE-cadherin1-31/mVE-cadherin32-739 (1-31 h/m), hVE-cadherin1-56/mVE-cadherin57-739 (1-56 h/m), and the mVE-cadherin1-56/hVE-cadherin57-737 (1-56 m/h) chimeric molecules tested. (B) Binding activity of BV13 and 10G4 on 293 cells transfected with the 1-31 h/m, 1-56 h/m, and 1-56 m/h chimeric VE-cadherin proteins, as compared to mock-transfected, and wild-type murine (mWT) and human wild-type (hWT) VE-cadherin, as measured by flow cytometry. Cells were stained with BV13 and 10G4 followed by goat anti–rat-PE secondary antibody to determine relative MFI by flow cytometry analysis (n = 3); error bars represent the standard deviation.
Identification of the disruptive mAb-binding domain. (A) Diagram illustrates the hVE-cadherin1-31/mVE-cadherin32-739 (1-31 h/m), hVE-cadherin1-56/mVE-cadherin57-739 (1-56 h/m), and the mVE-cadherin1-56/hVE-cadherin57-737 (1-56 m/h) chimeric molecules tested. (B) Binding activity of BV13 and 10G4 on 293 cells transfected with the 1-31 h/m, 1-56 h/m, and 1-56 m/h chimeric VE-cadherin proteins, as compared to mock-transfected, and wild-type murine (mWT) and human wild-type (hWT) VE-cadherin, as measured by flow cytometry. Cells were stained with BV13 and 10G4 followed by goat anti–rat-PE secondary antibody to determine relative MFI by flow cytometry analysis (n = 3); error bars represent the standard deviation.
The disruptive mAbs target an overlapping epitope. (A) Alignment of the murine and human VE-cadherin amino acid sequence up to position 56. Differences in the amino acid sequence are underlined and in boldface. Shown are the murine amino acids sequences 15-20, 27-31, 36-38, 41-42, 45-51, and 54-56 that were substituted with corresponding human sequence to generate the human-murine VE-cadherin chimeras (shown in brackets). (B) Binding activity of BV13 and 10G4 on 293 cells transfected with the VE-cadherin chimeras, as compared to mock-transfected and mWT VE-cadherin, as measured by flow cytometry. Cells were stained with BV13 and 10G4 followed by goat anti–rat-PE secondary antibody to determine relative MFI by flow cytometry analysis (n = 3); error bars represent the standard deviation.
The disruptive mAbs target an overlapping epitope. (A) Alignment of the murine and human VE-cadherin amino acid sequence up to position 56. Differences in the amino acid sequence are underlined and in boldface. Shown are the murine amino acids sequences 15-20, 27-31, 36-38, 41-42, 45-51, and 54-56 that were substituted with corresponding human sequence to generate the human-murine VE-cadherin chimeras (shown in brackets). (B) Binding activity of BV13 and 10G4 on 293 cells transfected with the VE-cadherin chimeras, as compared to mock-transfected and mWT VE-cadherin, as measured by flow cytometry. Cells were stained with BV13 and 10G4 followed by goat anti–rat-PE secondary antibody to determine relative MFI by flow cytometry analysis (n = 3); error bars represent the standard deviation.
Identification of amino acid residues required for adherens junction formation
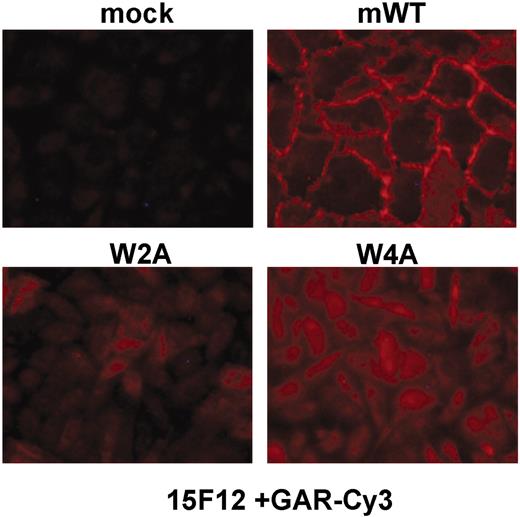
The classic cadherins maintain a conserved tryptophan at position 2 (W2) of the mature protein. W2 has been shown previously in N-, E-, and C-cadherin to be required for adhesion and the formation of adherens junctions.9,39-41 Unlike type I classic cadherins, type II classic cadherins maintain a second conserved tryptophan at position 4 (W4). The roles of W2 and W4 in VE-cadherin–mediated adhesion have not previously been established. To test the role of W2 and W4 in VE-cadherin–mediated adhesion, we generated stable cell lines expressing mWT, and the alanine mutants W2A and W4A of mVE-cadherin. We tested their ability to form adherens junctions by assessing VE-cadherin clustering at the cell-cell contacts. Although all cell lines expressed similar levels of VE-cadherin on their surface as determined by flow cytometry (data not shown), only cells expressing the wild-type molecule demonstrated distinct staining typical of VE-cadherin clustering at adherens junctions (Figure 5), indicating that either W2A or W4A mutations are deleterious to adherens junction formation. Therefore, we conclude that both the W2 and W4 residues are required for VE-cadherin function as a homotypic adhesion molecule. The ability of E4G10 to inhibit trans-dimerization may therefore be a function of its binding to a sequence adjacent to and including W4, thereby preventing W4 interaction with the hydrophobic pocket on the partner VE-cadherin molecule.
VE-cadherin homology modeling
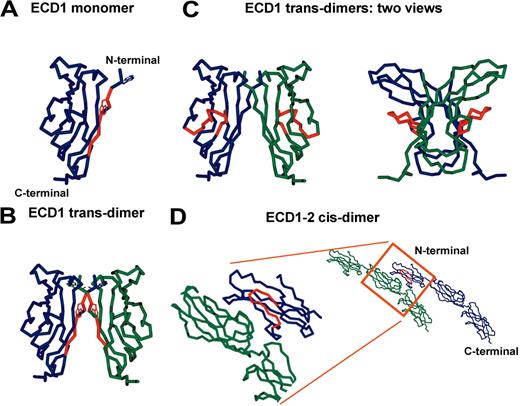
Similarities between known classic cadherin crystal structures allowed us to generate a 3-dimensional homology model of VE-cadherin extracellular domains (ECD1-ECD5) based on sequence alignments with C-cadherin.9 From this we identified the nondisruptive (amino acids 4, 5, 7-10) and disruptive (amino acids 45-56) antibody-binding epitopes on VE-cadherin in relation to the regions of VE-cadherin that appear involved in both trans- and cis-interactions (Figure 6). Our homology model predicts the nondisruptive epitope of E4G10 is accessible on the monomer form of VE-cadherin (Figure 6A), but becomes inaccessible on transdimerization (Figure 6B). In contrast, the disruptive epitope targeted by both BV13 and 10G4 appears accessible to antibody binding on VE-cadherin trans-dimers (Figure 6C), as well as the cis-interacting molecules (Figure 6D). Therefore, E4G10 demonstrates in vivo antiangiogenic activity because it can prevent the interaction of VE-cadherin monomers and is not disruptive because the epitope is inaccessible on dimerization, whereas BV13 and 10G4 are able to disrupt existing vasculature in vivo because their epitope is exposed regardless of whether or not VE-cadherin is engaged in homophilic interactions.
Conserved tryptophans are required for adherens junction formation. (A) CHO cells stably expressing wild-type (mWT), W2A, and W4A mVE-cadherin, along with mock-transduced cells, were examined for the VE-cadherin clustering at adherens junctions by immunohistochemistry (mAb 15F12 + goat anti–rat-Cy3 IgG).
Conserved tryptophans are required for adherens junction formation. (A) CHO cells stably expressing wild-type (mWT), W2A, and W4A mVE-cadherin, along with mock-transduced cells, were examined for the VE-cadherin clustering at adherens junctions by immunohistochemistry (mAb 15F12 + goat anti–rat-Cy3 IgG).
Discussion
Considerable understanding of the molecular mechanism of cell adhesion by type I classic cadherins has been achieved in recent years, mainly through extensive structural and functional studies including high-resolution x-ray7,9,39 and nuclear magnetic resonance (NMR) structures.40 Cadherin-mediated homophilic trans-adhesion requires the N-terminal domains (ECD1) of interacting molecules from adjacent cells. The trans-adhesive interface of E-, N-, and C-cadherin maintains a conserved tryptophan residue (W2) essential for dimerization, which inserts into a hydrophobic pocket formed within the ECD1 domain of the partner cadherin molecule on the opposing cell.3-5,13
Our findings suggest that the trans-interactions of type II cadherins in adherens junctions are similar to those of type I cadherins. In support of this view, an electron microscopy study of a 5-ECD VE-cadherin fusion protein revealed dimer formation mediated by the N-terminal domain, in a fashion essentially identical to that seen for the type I E-cadherin.42 Recently, other cadherin superfamily members, the desmosomal cadherins, have been shown by electron tomography to use adhesive interactions involving ECD1 in a manner analogous to that observed with type I cadherins.43 Our VE-cadherin studies, together with those reported for type I and desmosomal cadherins, demonstrate a dominant role for ECD1 in cadherin-mediated adhesion. The similarities between type I and II cadherins is further evident in our studies that demonstrate type II cadherins, like type I cadherins, require conserved N-terminal tryptophans for adhesion. However, type II cadherins appear distinct from type I cadherins in that they maintain 2 conserved tryptophan residues, W2 and W4, which are required for adhesion, instead of W2 alone. Yet the overall mechanism of ECD1-mediated trans-adhesion appears very similar. Although our work demonstrates the importance of ECD1 in VE-cadherin function, it cannot exclude the involvement of ECD2-5, as others have reported.11-17,44 Our inability to identify blocking antibodies that target the other ECDs may simply be due to the limited number of mAbs studied.
To better understand the molecular basis for the disruptive versus nondisruptive mAb activities, we developed a 3-dimensional homology model of VE-cadherin based on C-cadherin. Similarities between known classic cadherin crystal structures allowed us to generate a model of mVE-cadherin based on the interactions of C-cadherin (Figure 6). From this model, we propose a hypothesis for the differences in activity between E4G10 and the disruptive mAbs BV13 and 10G4. Our homology model predicts that the epitope of E4G10, located at the immediate N-terminal domain of VE-cadherin, is accessible on the monomer form of the molecule as it exists on the nascent vasculature prior to trans-dimerization (Figure 6A). In support of this, crystallographic analysis has shown the corresponding N-terminal sequences of both C- and N-cadherin to be exposed in their monomeric conformations.18 In contrast to the monomer, at least part of the epitope is engaged in interactions on trans-dimer formation, and thus is masked on established vessels (Figure 6B). Therefore, the antibody E4G10 is antiangiogenic because it can prevent the functional interaction of VE-cadherins at a time when adherens junctions are formed in the process of neovascularization, and is not disruptive to existing vasculature because the epitope is inaccessible to the mAb.
In contrast, the disruptive antibodies BV13 and 10G4 both recognize epitopes comprised primarily of residues within amino acids 45-56 of mVE-cadherin. Homology modeling suggests that this sequence represents a well accessible surface loop in the first domain of VE-cadherin (Figure 6C). This epitope, although close to the E4G10 epitope, is located on a different face of ECD1, with homology modeling clearly suggesting that it does not take part in trans-dimer formation. The accessibility of the disruptive epitope is supported by earlier experiments that detected BV13 binding to both normal (lung, heart, kidney) and tumor vasculature in vivo, whereas E4G10 targeted only tumor vasculature.29 Therefore, we propose that BV13 and 10G4 are able to disrupt existing vasculature because residues 45-56 are exposed, regardless of whether or not VE-cadherin is engaged in adherens junctions. This epitope partially overlaps a previously identified structure on N-cadherin, termed the quasi-β-helix.45 We do not yet understand how targeting BV13 and 10G4 to residues within the surface loop of sequence 45-56 leads to destabilization of adherens junctions. It is possible that antibody binding to this site may cause changes in the conformation of ECD1, with detrimental effects on the stability of the VE-cadherin trans-dimer interface. Alternatively, because our homology model predicts the epitope of the disruptive mAbs is located in close proximity to the proposed cis-adhesive interface (Figure 6D), mAbs that bind this loop structure may interfere with the cis-interacting domains in such a way that they cannot maintain adhesion. If true, this would be evidence that the cis-interface is critical for stabilizing cadherin-mediated adhesion.
Homology model of VE-cadherin. Structural homology model of murine VE-cadherin ECDs based on the crystal structure of C-cadherin. The epitope of E4G10 (amino acids 4, 5, 7-10) is highlighted in red (A) on the monomer form of VE-cadherin ECD1, and (B) VE-cadherin ECD1 as a trans-dimer. (C) The epitope of BV13 and 10G4 (amino acid sequence 45-56) is highlighted in red on 2 different views of the VE-cadherin ECD1 trans-dimer. (D) The epitope of BV13 and 10G4 (amino acid sequence 45-56) is highlighted in red on cis-interacting VE-cadherin ECD1-2 molecules.
Homology model of VE-cadherin. Structural homology model of murine VE-cadherin ECDs based on the crystal structure of C-cadherin. The epitope of E4G10 (amino acids 4, 5, 7-10) is highlighted in red (A) on the monomer form of VE-cadherin ECD1, and (B) VE-cadherin ECD1 as a trans-dimer. (C) The epitope of BV13 and 10G4 (amino acid sequence 45-56) is highlighted in red on 2 different views of the VE-cadherin ECD1 trans-dimer. (D) The epitope of BV13 and 10G4 (amino acid sequence 45-56) is highlighted in red on cis-interacting VE-cadherin ECD1-2 molecules.
Because we relied on mutational analysis to delineate our mAb epitopes, we would like to emphasize our approach using chimeric VE-cadherin mutants. VE-cadherin is a complex protein that requires self-assembly into a multimeric complex for function. Therefore, we favored a method whereby all mutations were generated in a manner that would preserve structural integrity. Because type I and type II cadherins are thought to have essentially similar 3-dimensional structures, we reasoned that by replacing sequences of murine VE-cadherin with those of corresponding murine N-cadherin or human VE-cadherin sequences, we would minimize the potential for undesirable structural changes. In doing so, we were able to identify the mAb epitopes facilitated by the differential binding to the various chimeras.
In summary, our current findings suggest E4G10 recognizes an epitope on VE-cadherin that is only accessible on vascular endothelial cells prior to adherens junction formation and is not exposed in established adherens junctions. We therefore have identified a mAb epitope that is temporally exposed on a specific subset of the vasculature, namely, the neovasculature, yet the molecule itself is constitutively expressed throughout the entire vasculature. Therefore, this target region may represent an “angiogenic-epitope” and could potentially provide the basis for a novel approach to angiogenic-specific vascular targeting.
A number of advances have recently been made in immunoconjugate technology. As a result, several studies demonstrating effective antibody delivery of covalently linked or chelated toxic effector molecules for the treatment of cancer have been reported.46 We plan to determine if targeting the neovasculature with either radionuclide- or toxin-conjugated E4G10 results in increased tumor inhibition, as compared to what we have previously observed with the antibody alone.29
Prepublished online as Blood First Edition Paper, February 8, 2005; DOI 10.1182/blood-2005-01-0010.
Supported by research funding from ImClone Systems Incorporated (C.M., J.F.D., R.A., P.B., X.X., Z.Z., P.K., D.J.H., F.L., and P.B.). Several authors (C.M., J.F.D., P.B., Z.Z., P.K., D.J.H., and P.B.) are employed by and have declared a financial interest in a company (ImClone Systems Incorporated) whose potential product was studied in the present work. Three authors (R.A., X.X., and F.L.) were employed by ImClone Systems Incorporated at the time of this study.
An Inside Blood analysis of this article appears at the front of the issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Dr A. Vecchi for providing the H5V cells, and Bridget Finnerty, Dan Lu, and Rajiv Bassi for their excellent technical assistance.