Comment on Flomenberg et al, page 1923
The central question in selecting an unrelated stem cell donor is whether we should try to match for all HLA loci and at the highest typing resolution, ie, at the allele level.
Bone marrow transplantation became a clinical reality when it was realized that HLA matching was a conditio sine qua non. In family studies, serology can be used to identify the HLA haplotypes and can thus reliably select HLA-identical sibling donors. It was soon realized that matching unrelated donor-recipient pairs only for HLA-A, -B, and -DR by serology gave suboptimal results compared with HLA-identical siblings. Furthermore, selecting unrelated donors that were negative in the cytotoxic T-lymphocyte precursor test with the patient led to better transplantation outcome and made it clear that our typing and matching procedures had been inadequate.1 Many studies using DNA technology were started and a consensus was reached that one should attempt to find a donor who was matched at the allele level for HLA-A, -B, and -DRB1. Whether or not HLA-C, -DQ, or -DP should be included remained an open question and a matter of hot dispute.
In this issue of Blood, Flomenberg and colleagues provide convincing evidence that one should attempt to match at the allele level not only for HLA-A, -B, and -DRB1 but also for HLA-C. The authors studied 1874 donor-recipient pairs that had all been HLA typed at the allele level for HLA-A, -B, -C, -DR, -DQ, and -DP (pairs were selected from the National Marrow Donor Program files). To assess the effect of high-resolution matching at the different loci on transplantation outcome, engraftment, graft-versus-host disease, and mortality were used as end points.
In this large study, no significant evidence was found that matching for HLA-DQ and -DP matters. Because this can only be studied adequately in HLA-A, -B, -C, and -DRB1 identical siblings, this does not imply that they are of no importance.2
These findings are good news for about 60% of the patients with a relatively frequent high-resolution HLA phenotype, because high-resolution HLA matching will improve transplantation outcome. Obviously, though, this is bad news for the remaining patients.
The situation is especially grim for patients carrying HLA phenotypes with a low frequency in donor registries and cord blood banks. Of the more than 9 million unrelated stem cell donors worldwide (including cord blood units), 10% have an HLA low-resolution phenotype with a frequency of fewer than 1 per million donors, so-called unique phenotypes.3 Assuming that the HLA phenotype frequency distribution is about the same in patients and donors, it will be near impossible to find an optimally matched donor for such patients. The situation for high-resolution phenotypes will be obviously much worse. Extrapolating from available evidence, more than 20% of the patients will have such a unique high-resolution HLA-A, -B, -C, -DRB1 phenotype. Worse, we are only discussing the situation for patients with West European genetic background!
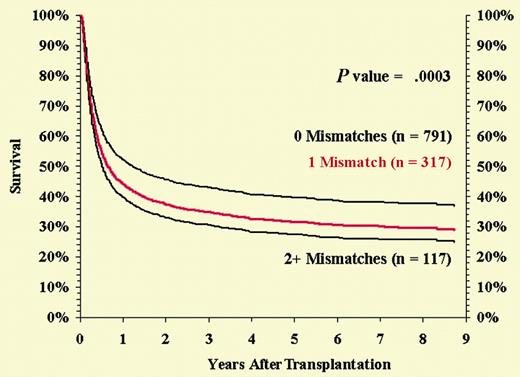
Flomenberg et al recognize this problem and emphasize that “adding more typing information to the process of donor selection should not be used to exclude more patients from the potentially curative benefits of this therapy.” They also suggest that if larger numbers of transplantations could be analyzed it might be possible to identify mismatches that are more “permissible” than others.FIG1
Risk-adjusted survival among HLA-A, -B serologic, and -DRB1 allele-matched pairs by number of class I loci mismatched at the allele level. See the complete figure in the article beginning on page 1923.
Risk-adjusted survival among HLA-A, -B serologic, and -DRB1 allele-matched pairs by number of class I loci mismatched at the allele level. See the complete figure in the article beginning on page 1923.
Organ-sharing organizations have collected important information on permissible antigen mismatches in kidney transplantation. This was only possible because they were able to analyze files of 10 000 or more transplantations.4 With more than 5000 transplantations per year worldwide, it ought to be possible to obtain similarly large files of stem cell transplantations with unrelated donors. Such files might provide more information not only on permissible mismatches but also on other parameters such as conditioning protocols vis-à-vis HLA mismatching, the role of natural killer (NK) cells, the use of cord blood in adult patients, and so on.5
This implies, however, a global effort. The study by Flomenberg et al is a good start and the forthcoming results of the 13th Histocompatibility Workshop are a good follow-up, but they regard only a fraction of the stem cell transplantations performed annually worldwide. Because stem cell transplantation poses such a high mortality risk to the patients and such a great expense to the community making it available, we have the moral obligation to attempt to realize a uniform global follow-up of stem cell transplantations. ▪