Abstract
CCAAT enhancer binding protein-α (C/EBPα) inhibits proliferation in multiple cell types; therefore, we evaluated whether C/EBPα-deficient hematopoietic progenitor cells (HPCs) have an increased proliferative potential in vitro and in vivo. In this study we demonstrate that C/EBPα-/- fetal liver (FL) progenitors are hyperproliferative, show decreased differentiation potential, and show increased self-renewal capacity in response to hematopoietic growth factors (HGFs). There are fewer committed bipotential progenitors in C/EBPα-/- FL, whereas multipotential progenitors are unaffected. HGF-dependent progenitor cell lines can be derived by directly culturing C/EBPα-/- FL cells in vitro Hyperproliferative spleen colonies and myelodysplastic syndrome (MDS) are observed in mice reconstituted with C/EBPα-/- FL cells, indicating progenitor hyperproliferation in vitro and in vivo. C/EBPα-/- FL lacked macrophage progenitors in vitro and had impaired ability to generate macrophages in vivo. These findings show that C/EBPα deficiency results in hyperproliferation of HPCs and a block in the ability of multipotential progenitors to differentiate into bipotential granulocyte/macrophage progenitors and their progeny. (Blood. 2004; 104:1639-1647)
Introduction
Hematopoiesis is a complex cellular process whereby short-lived mature blood cells are continuously replenished by a limited number of hematopoietic stem cells (HSCs) through rapidly dividing transit cell populations.1-4 This process is regulated, in part, by the hematopoietic microenvironment (stromal cells and extracellular matrix) and hematopoietic growth factor (HGF) that bind to cell surface receptors, which activate signal transduction pathways to either positively or negatively regulate transcription factors.5 Transcription factors are essential for development and affect hematopoiesis by regulating the expression of HGF, HGF receptors, other transcription factors, and lineage-specific genes.6-9 For example, one member of the CCAAT enhancer binding protein (C/EBP) transcription factor family, C/EBPα, is critical for myeloid development in vitro and in vivo.10,11 Forced expression of C/EBPα in some hematopoietic progenitor cell (HPC) lines induces granulocyte differentiation, in certain cases at the expense of macrophage differentiation.12,13 Furthermore, neutrophil maturation is arrested in C/EBPα-/- mice, because in part of the absence of granulocyte colony-stimulating factor (G-CSF) and interleukin 6 (IL-6) receptor expression, which are required to promote normal granulocyte maturation.14
In addition to regulating neutrophil maturation, C/EBPα is also critical for the function of other cell types and regulates the expression of genes that are required for lipid accumulation and glycogen storage.15-17 In this regard, C/EBPα-/- mice die shortly after birth as a result of hypoglycemia. Therefore, in the studies described in this report, we used FL cells to evaluate the role of C/EBPα in hematopoietic development. C/EBPα has been shown to affect the growth and proliferation of a number of cell types in vitro and in vivo. For example, overexpression of C/EBPα inhibits the growth of hepatocyte, adipocyte, and myeloid progenitor cell lines.18-20 Conversely, fetal hepatocytes from C/EBPα-/- mice display increased proliferative capacity in vitro,21 and C/EBPα-/- FL resembles that of regenerating liver with increased levels of c-myc, c-jun, and proliferating cell nuclear antigen (PCNA) gene expression.17,22 Also, histologic analysis of C/EBPα-/- lung tissue indicates a hyperproliferation of type II pneumocytes. Other C/EBP family members have also been implicated in negative regulation of cell growth. For example, C/EBPα-/- mice develop myeloid hyperplasia,23 C/EBPα-/- mice show a lymphoproliferative disorder after 16 weeks,24 and enforced expression of C/EBPα can promote growth arrest of mammary epithelial cells.
It has recently been proposed that loss of C/EBPα expression or function may contribute to the differentiation block, enhanced proliferation, and development of some acute myelogenous leukemias (AMLs).25 For example, the expression of the AML-ETO fusion protein in some patients inhibits C/EBPα transcription, whereas the expression of bcr-abl in other patients prevents C/EBPα translation.26-28 Also, heterozygous mutations at the C/EBPα locus, which generate dominant-negative forms of C/EBPα, have been observed in patients with AML.29-31 Although these data support the hypothesis that loss of C/EBPα may lead to the generation of AML, it is not clear what additional genetic events are required for loss of growth control during leukemogenesis in these patients. These observations support the hypothesis that C/EBPα and other family members play critical roles in regulating the balance between proliferation and growth arrest/differentiation in hematopoietic progenitors and other cell types. Therefore, we asked whether C/EBPα might play a role in regulating the proliferation of primitive and more committed HPCs in vitro and in vivo. To address this question, we determined whether progenitors from C/EBPα-/- fetal liver (FL) showed increased proliferative potential in vitro and in vivo. In addition, we undertook a detailed progenitor analysis to more precisely define the block to myeloid differentiation imposed by C/EBPα deficiency. Furthermore, we examined the maturation of C/EBPα-/- FL cells transplanted in vivo to determine whether the defects were intrinsic to HPCs or the hematopoietic microenvironment.
Materials and methods
Mice
The C/EBPα+/- strain used in these experiments is on a mixed background of 129sv and C57Bl/6.32 For transplantation studies, the C/EBPα+/- mice were backcrossed 9 generations onto a pure C57Bl/6 background (Ly-5.1). Both strains were maintained at the animal production area at the National Cancer Institute at Frederick (Frederick, MD). Animal care was provided in accordance with the procedures outlined in the “Guide for Care and Use of Laboratory Animals.”33 Mice were maintained in a pathogen-free environment.
Cells
FL was obtained from day 16 to 17 embryos (E16-17) from C/EBPα+/- pregnant mothers. Peripheral blood (PB) mononuclear cells and bone marrow cell (BMC) suspensions were incubated with a mixture of monoclonal antibodies in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA) for fluorescence-activated cell sorting (FACS) analysis. Peritoneal exudate cells (PECs) were harvested by lavage with 10 mL PBS containing 5 mM EDTA (ethylenediaminetetraacetic acid) just before (resident PEC) and 72 hours after (elicited PEC) injection of 2 mL, 3% thioglycollate medium (DIFCO, Detroit, MI).
Soft agar colony formation assay
FL cells from C/EBPα-/-, C/EBPα+/-, and C/EBPα+/+ mice were plated in cIMDM (Iscoves modified Dulbecco medium containing 10% FCS, 2 mM l-glutamine, 1% penicillin-streptomycin) in the presence or absence of HGFs as previously described.34
Northern blot analysis and RNase protection assays
Total RNA was purified with use of RNeasy (Qiagen, Chatsworth, CA) as outlined by the manufacturer. Separation of RNA samples (10-15 μg) by electrophoresis was performed on 1% agarose, 5.2% formaldehyde (37% solution), 1 × MOPS gels and then transferred to nylon membrane (MSI, Westboro, MA) as previously described.35 Macrophage colony-stimulating factor receptor (M-CSFR; gift of Dr Larry Rorschneider) and glyceraldehydes-3-phosphate dehydrogenase (GAPDH; Clontech, Palo Alto, CA) cDNA probes were labeled with random primers with use of Prime-It II (Stratagene, La Jolla, CA) following the manufacturers' instructions. For ribonuclease protection assay (RPA), total RNA was extracted from E16 to 17 day FL cells and the IL-3-dependent C/EBPα-/- cell line with use of Trizol (Invitrogen, Carlsbad, CA). A probe set containing murine granulocyte-macrophage colony-stimulating factor receptor α (GM-CSFRα), IL-6R, and gp130 (BD-Pharmingen, San Diego, CA) was labeled with 32P-UTP (uridine triphosphate) (Amersham, Arlington Heights, IL) with use of the Riboquant In Vitro assay kit (BD-Pharmingen). Protection assays were performed on 10 μg RNA with use of the Riboquant RNase Protection Assay kit (BD-Pharmingen) as recommended by the supplier. GM-CSFRα and IL-6R transcripts were quantitated with use of a Storm 860 phosphorimager (Molecular Dynamics, Piscataway, NJ) and normalized to GAPDH.
FL transplantation and spleen colony-forming unit (CFU-S) assay
E16 to E17 day C/EBPα-/-, C/EBPα+/-, and C/EBPα+/+ FL cells (Ly-5.1) were combined with normal BMCs (Ly-5.2) to radioprotect lethally irradiated Ly-5.2 recipient mice. Mice were exposed to 10 Gy total body (137Cesium irradiation) 3 hours prior to the intravenous injection of FL cells. Reconstitution was evaluated by FACS analysis with use of antibodies that recognize host cells (Ly-5.2) and donor-derived cells (Ly-5.1), and antibodies that recognize granulocytes (Gr-1, RB6-8C5), macrophages (Mac-1 and F4.80), B cells (B220), or T cells (CD3α) and isotype-matched control antibodies (Pharmingen) (F4.80 from e-Bioscience, San Diego, CA).
Proliferation assays
To measure proliferation, cells were washed 3 times in cIMDM and plated at 1 × 104 cells/100 μL in microtiter plates in triplicate with the indicated HGF. Cells were incubated for 48 hours and pulsed with 1 μCi (0.037 MBq) 3H-thymidine per well (6.7 Ci/mmol/L [24.79 × 1010 Bq]; NEN, Boston, MA) for the last 6 to 8 hours of incubation and then harvested (Tomtech Harvester 96; Tomtech, Orange, CT) onto glass-fiber filter paper (Filtermat A; Wallac Oy, Turku, Finland) for liquid scintillation counting.
Flow cytometry
The cells were washed and resuspended in Hanks balanced salt solution (HBSS) containing 0.02% BSA at a density of 1 × 107 cells/mL. The cells were incubated with fluorochrome-labeled c-Kit, Sca-1, CD-34, Ter-119, Gr-1, Mac-1, F4.80, Moma-1, B220, CD-3, CD-4 (Pharmingen) at 1 μg/L × 106 cells/100 μL in the same buffer for 30 minutes at 4°C, washed, and resuspended in the same buffer for FACS analysis.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed with use of a double-stranded oligonucleotide probe containing the consensus C/EBP binding site, as described.36 For supershift assays, the nuclear extract was preincubated with 2 μL rabbit antiserum at 4°C for 30 minutes. Antibodies against C/EBP proteins were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), except for the C/EBPγ antiserum, which has been described previously.36
Results
Effect of HGFs on the proliferation of C/EBPα-/- FL cells in vitro
To determine whether primitive HPCs that respond to multiple HGF combinations and committed HPCs that respond to single HGF from C/EBPα-/- mice were hyperproliferative, FL cells were plated in soft agar colony assays (culture colony-forming unit [CFU-C]) in the presence of HGF. As expected,11 C/EBPα-/- FL cells did not proliferate in response to G-CSF, whereas C/EBPα+/+ and C/EBPα+/- FL cells showed a normal G-CSF response (Figure 1A). In contrast to what has been previously observed,14 the number of IL-3- and GM-CSF-responsive C/EBPα-/- CFU-Cs were decreased by 50% and 80%, respectively, compared with the controls. Furthermore, there was a 95% decrease in the number of M-CSF-responsive CFU-Cs, and an absence of SCF-responsive CFU-Cs in C/EBPα-/- FL. Interestingly, little or no effect on the number of more primitive SCF/IL-3-responsive CFU-Cs was observed in the C/EBPα-/- FL cell population.
Effect of hematopoietic growth factors (HGFs) on the growth of C/EBPα-/- fetal liver (FL) cells in vitro. (A) FL cells were obtained from E16-E17 C/EBPα-/- mice and seeded into soft agar colony assays at 5 × 104 cells/mL in the presence or absence of 50 ng/mL human G-CSF (Amgen, Thousand Oaks, CA), 100 ng/mL M-CSF, 100 ng/mL murine SCF, 50 ng/mL murine GM-CSF, or 30 ng/mL murine IL-3 (Peprotech, Rocky Hill, NJ). (B) Bright field photomicrographs (× 16) of the 35-mm gridded (2 mm × 2 mm) plates were taken after 7 days. Dishes were incubated in a fully humidified atmosphere at 37°C, 5% CO2, and then scored for colony formation after 7 to 10 days (culture colony-forming unit [CFU-C]). The results are presented as the mean number of colonies plus or minus the standard error, and are representative of at least 3 separate experiments.
Effect of hematopoietic growth factors (HGFs) on the growth of C/EBPα-/- fetal liver (FL) cells in vitro. (A) FL cells were obtained from E16-E17 C/EBPα-/- mice and seeded into soft agar colony assays at 5 × 104 cells/mL in the presence or absence of 50 ng/mL human G-CSF (Amgen, Thousand Oaks, CA), 100 ng/mL M-CSF, 100 ng/mL murine SCF, 50 ng/mL murine GM-CSF, or 30 ng/mL murine IL-3 (Peprotech, Rocky Hill, NJ). (B) Bright field photomicrographs (× 16) of the 35-mm gridded (2 mm × 2 mm) plates were taken after 7 days. Dishes were incubated in a fully humidified atmosphere at 37°C, 5% CO2, and then scored for colony formation after 7 to 10 days (culture colony-forming unit [CFU-C]). The results are presented as the mean number of colonies plus or minus the standard error, and are representative of at least 3 separate experiments.
In addition to the number of CFU-Cs, the size of the colonies can be readily evaluated in soft agar. As expected, most of the colonies present in C/EBPα+/+ FL cultures containing IL-3 or GM-CSF consisted of committed progenitor cells that were less than 1 mm in diameter (Figure 1B). In contrast, most of the IL-3- or GM-CSF-responsive CFU-Cs from C/EBPα-/- FL were large colonies more than 1 mm in diameter and frequently as large as 2 mm, a characteristic of more primitive progenitors with a higher proliferative potential (Figure 1B). Furthermore, colonies more than 4 to 6 mm in diameter were observed when C/EBPα-/- FL cells were grown in multi-HGF conditions (SCF plus IL-3). C/EBPα+/+ FL cells in liquid cultures contained few mitotic cells (cytocentrifuge preparations), and the number of cells in S phase, as determined by propidium iodide staining, was 2% for cells cultured in GM-CSF or IL-3 cultures. In comparison, cytospins of C/EBPα-/- FL cell cultures contained many mitotic cells, and the number of cells in S phase was 8.0% ± 2.4% for GM-CSF and 12.1% ± 3% for IL-3 cultures (data not shown). Taken together, C/EBPα-/- FL lack more mature unipotential HPCs that respond to G-CSF, M-CSF, and SCF and have reduced numbers of committed HPCs that respond to GM-CSF or IL-3. However, the remaining GM-CSF-, IL-3-, and IL-3/SCF-responsive HPCs from C/EBPα-/- FL are hyperproliferative.
HGF receptor expression on C/EBPα-/- FL cells
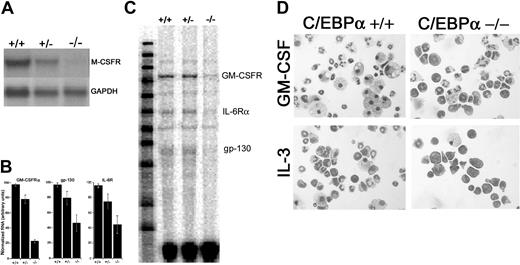
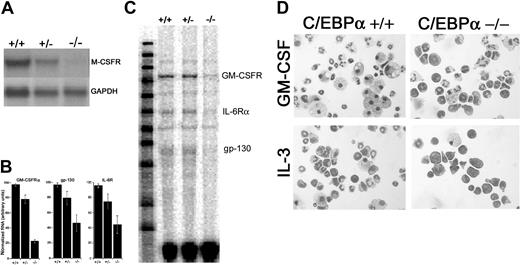
The absence of neutrophils in C/EBPα-/- mice is thought to be due, in part, to the lack of G-CSF and IL-6 receptor expression.14 Therefore, we sought to determine whether decreased M-CSF and GM-CSF receptor expression could account for the absence of M-CSF-responsive and decreased GM-CSF-responsive progenitor cells. RNA was obtained from E16 to E17 day C/EBPα-/-, C/EBPα+/+, and C/EBPα-/+ FL and analyzed for M-CSFR expression by Northern blot hybridization (Figure 2A). Little or no M-CSFR RNA was detected in the C/EBPα-/- FL by Northern blot analysis, whereas high levels were observed in C/EBPα+/+ and intermediate levels in C/EBPα+/- FL. In addition, analysis of GM-CSFRα mRNA by RNase protection assay demonstrates that GM-CSFRα expression was reduced by roughly 80% in C/EBPα-/- FL (Figure 2B-C) compared with C/EBPα+/+ and C/EBPα+/- controls. In agreement with previous findings,14 we observed decreased IL-6Rα and gp130 levels in C/EBPα-/- FL compared with C/EBPα+/+ and C/EBPα+/- controls in RNase protection assays (Figure 2B-C). Taken together, the absence of M-CSF receptor and decreased expression of GM-CSF correlate with decreased growth of C/EBPα-/- FL cells in response to M-CSF and GM-CSF in vitro.
Expression of HGF receptors in C/EBPα-/- FL cells and morphology of C/EBPα-/- FL cells grown in IL-3 and GM-CSF. RNA was extracted from E16 to 17 C/EBPα+/+, C/EBPα+/-, and C/EBPα-/- FL and analyzed by Northern blot for the expression of M-CSFR and GADPH (A) or by RPA for GM-CSFRα, IL-6R, and gp130 according to the procedures outlined in “Materials and methods” (C). RPA signals were quantitated, relative to controls, by scanning densitometry (B). FL cells were harvested from E16 to 17 C/EBPα+/+ and C/EBPα-/- mice seeded into liquid cultures at 2 × 105 cells/mL in cIMDM. Cells were removed from the cultures after 7 days of incubation (37°C) in the presence of IL-3 or GM-CSF, cytocentrifuged, and stained with Giemsa. Bright field photomicrographs were taken with a Leica DMLB microscope at a magnification of × 400 (D).
Expression of HGF receptors in C/EBPα-/- FL cells and morphology of C/EBPα-/- FL cells grown in IL-3 and GM-CSF. RNA was extracted from E16 to 17 C/EBPα+/+, C/EBPα+/-, and C/EBPα-/- FL and analyzed by Northern blot for the expression of M-CSFR and GADPH (A) or by RPA for GM-CSFRα, IL-6R, and gp130 according to the procedures outlined in “Materials and methods” (C). RPA signals were quantitated, relative to controls, by scanning densitometry (B). FL cells were harvested from E16 to 17 C/EBPα+/+ and C/EBPα-/- mice seeded into liquid cultures at 2 × 105 cells/mL in cIMDM. Cells were removed from the cultures after 7 days of incubation (37°C) in the presence of IL-3 or GM-CSF, cytocentrifuged, and stained with Giemsa. Bright field photomicrographs were taken with a Leica DMLB microscope at a magnification of × 400 (D).
We would expect that overexpression of C/EBPα in C/EBPα-/- FL would result in increased M-CSF response and macrophage development. Therefore, we infected C/EBPα-/- FL cells with retroviral vectors that express C/EBPα and measured macrophage development in soft agar in response to M-CSF or M-CSF/SCF. M-CSF- and M-CSF/SCF-induced macrophage colony formation in C/EBPα-/- FL cells infected with C/EBPα-expressing retroviral vectors (Table 1).
Effect of HGF on the differentiation of C/EBPα-/- FL cells in vitro
Both GM-CSF and IL-3 promote the proliferation and differentiation of granulocytes and macrophages; therefore, we sought to determine the identity of the cells that were present in the hyperproliferating soft agar colonies from C/EBPα-/- FL cells. As expected, GM-CSF and IL-3 promoted the terminal differentiation of macrophages and granulocytes from C/EBPα+/+ FL cells after 7 days in culture (Figure 2D); IL-3 also induced mast cell differentiation in these cultures. In comparison, there was an increase in the number of primitive myeloid cells and a decrease in the number of differentiated granulocytes and macrophages in C/EBPα-/- FL cultures. Differential cell counts of cytospins showed that 65% of the C/EBPα-/- FL cells cultured in IL-3 are primitive myeloblasts and promyelocytes compared with 15% for the C/EBPα+/+ FL control cell cultures. In addition, only 3% of the cells in C/EBPα-/- FL cultures were a more differentiated band and segmented neutrophils compared with 45% for the C/EBPα+/+ FL. Similar results were obtained for FL cells cultured in GM-CSF. Thus, C/EBPα-/- FL progenitor cells show both increased proliferative potential and reduced differentiation in response to IL-3 or GM-CSF.
Increased plating potential of C/EBPα-/- FL cells
C/EBPα-/- FL cells grown in the presence of IL-3 or GM-CSF contain increased numbers of primitive myeloid progenitor cells. Therefore, we asked whether C/EBPα-/- FL cells from hyperproliferative primary colonies could give rise to secondary CFU-Cs in replating assays. To examine this, we removed FL cells that had been cultured for 7 days in IL-3 or GM-CSF and plated them in soft agar colony assays. As expected, control C/EBPα+/+ FL cells grown for 7 days in IL-3 or GM-CSF were unable to give rise to secondary CFU-Cs because these factors promote terminal differentiation and cell growth arrest during the initial culture period (Table 2). By contrast, C/EBPα-/- FL cells grown in IL-3 or GM-CSF and plated in the same cytokines generated large numbers of secondary CFU-Cs, indicating that progenitor cells in these cultures are capable of self-renewal. Interestingly, C/EBPα-/+ FL showed a haplo-insufficient phenotype, with a plating potential that was greater than that of C/EBPα+/+ FL cells but less than C/EBPα-/- FL cells, suggesting that partial loss of C/EBPα can also affect HPC proliferation.
Establishment of HGF-dependent cell lines from C/EBPα-/- FL
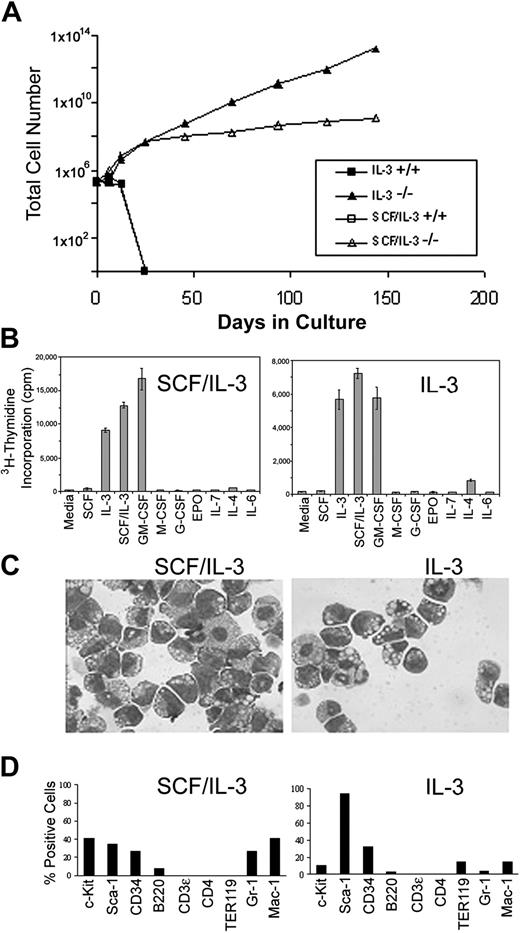
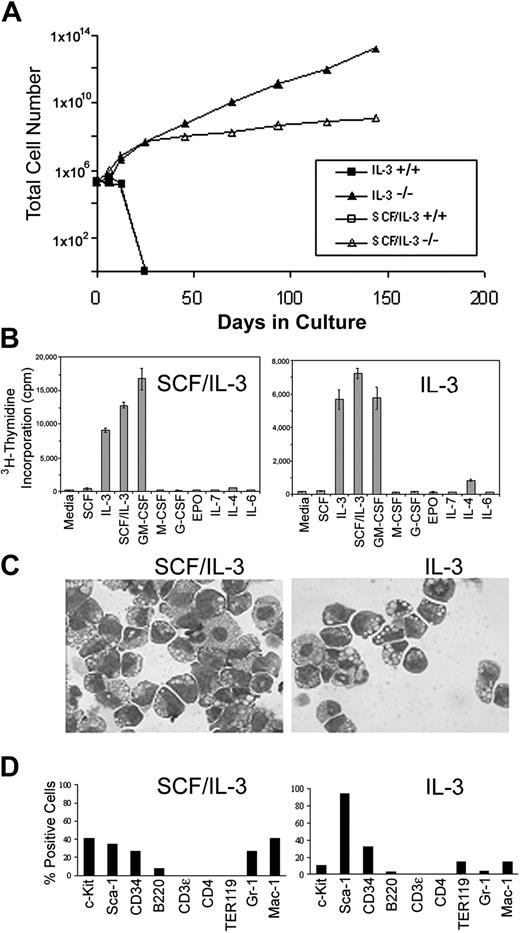
To determine the extent of C/EBPα-/- FL cell self-renewal and proliferation in vitro, C/EBPα-/- and C/EBPα+/+ FL cells were plated in liquid cultures in the presence or absence of IL-3 or IL-3 plus SCF, and the numbers of viable cells were evaluated over time. As expected, these growth factors promoted the growth and subsequent terminal differentiation of granulocytes, macrophages, and mast cells from C/EBPα+/+ FL, and by 3 weeks most of the cells had ceased to proliferate (Figure 3A). Mast cells slowly proliferated and persisted in these cultures for 2 to 3 months (data not shown). In contrast, IL-3 and IL-3 plus SCF promoted the rapid and continued proliferation and expansion of C/EBPα-/- FL cells. The C/EBPα-/- FL cultures have been maintained for more than 1 year in culture without a reduction in proliferation.
Establishment of SCF/IL-3- and IL-3-dependent cells from C/EBPα-/- FL in vitro. (A) C/EBPα+/+ and C/EBPα-/- FL cells were plated in cIMDM at 2 × 105 cells/mL in the presence or absence of IL-3 or IL-3 plus SCF. Total viable cell numbers were determined over time by trypan blue exclusion and hemocytometer counting. (B) SCF/IL-3-2 and IL-3B cells were plated in proliferation assays in the presence or absence of 100 ng/mL SCF, 30 ng/mL IL-3, 50 ng/mL GM-CSF, 100 ng/mL human IL-7 (Peprotech), 100 ng/mL human IL-6 (Peprotech), 5 U/mL human erythropoietin (EPO), 50 ng/mL G-CSF (Amgen), 100 ng/mL human M-CSF (Peprotech), 500 U/mL murine interferon α (IFNα; Biosource, Camarillo, CA), or 50 mg/mL IL-4 (Peprotech) according to the procedures outlined in “Materials and methods.” (C) Cytocentrifuge preparations of cell clones (SCF/IL-3-2 and IL-3B) derived from these cultures were stained with Giemsa, and bright field photomicrographs were obtained at magnification × 400. (D) The expression of lineage-specific cell surface antigens was measured with use of fluorochrome-labeled antibodies and flow cytometry according to the procedures outlined in “Materials and methods.” Percentage positive for each marker was obtained by subtracting the background fluorescence of an isotype matched control.
Establishment of SCF/IL-3- and IL-3-dependent cells from C/EBPα-/- FL in vitro. (A) C/EBPα+/+ and C/EBPα-/- FL cells were plated in cIMDM at 2 × 105 cells/mL in the presence or absence of IL-3 or IL-3 plus SCF. Total viable cell numbers were determined over time by trypan blue exclusion and hemocytometer counting. (B) SCF/IL-3-2 and IL-3B cells were plated in proliferation assays in the presence or absence of 100 ng/mL SCF, 30 ng/mL IL-3, 50 ng/mL GM-CSF, 100 ng/mL human IL-7 (Peprotech), 100 ng/mL human IL-6 (Peprotech), 5 U/mL human erythropoietin (EPO), 50 ng/mL G-CSF (Amgen), 100 ng/mL human M-CSF (Peprotech), 500 U/mL murine interferon α (IFNα; Biosource, Camarillo, CA), or 50 mg/mL IL-4 (Peprotech) according to the procedures outlined in “Materials and methods.” (C) Cytocentrifuge preparations of cell clones (SCF/IL-3-2 and IL-3B) derived from these cultures were stained with Giemsa, and bright field photomicrographs were obtained at magnification × 400. (D) The expression of lineage-specific cell surface antigens was measured with use of fluorochrome-labeled antibodies and flow cytometry according to the procedures outlined in “Materials and methods.” Percentage positive for each marker was obtained by subtracting the background fluorescence of an isotype matched control.
To evaluate the maturational state of the C/EBPα-/- FL cell lines, single cell clones were established from bulk cultures in SCF plus IL-3 or IL-3 alone and then analyzed for (1) response to HGF, (2) morphology by Giemsa staining, and (3) lineage analysis by cell surface antigen expression and flow cytometry. Cloned cell lines established from C/EBPα-/- FL cells grown in SCF plus IL-3 (SCF/IL-3-2 and SCF/IL-3-1B) (not shown) and IL-3 (IL-3-3B) were further analyzed. Both SCF/IL-3-2 and IL-3B proliferated in response to IL-3, GM-CSF, and SCF plus IL-3 but did not proliferate in response to medium alone or the other HGFs (G-CSF, EPO, IL-7, IL-4, and IL-6) (Figure 3B). Morphologic analysis showed that the cloned cell lines consisted of blast cells, promyelocytes, and myelocytes with an occasional appearance of more differentiated cells (Figure 3C). In agreement with this, cell surface staining showed that the cell lines expressed antigens present on primitive progenitors, including c-Kit, Sca-1, and CD-34. In addition, they expressed antigens found on macrophages, including Mac-1 (F4/80, data not shown) (low levels of Mac-1 expressed on stem cells), low levels of Gr-1 (granulocytes) and B220 (B cells) and, low or no expression of Ter-119 (erythroid) and CD3 or CD4 (T-cell antigens) (Figure 3D). Taken together, loss of C/EBPα gene expression in HPCs results in hyperproliferation with a concomitant block in differentiation that leads to establishment of HGF-dependent progenitor cell lines in vitro. Furthermore, these cell lines resemble HPCs blocked at the common myeloid progenitor (CMP) and granulocyte/macrophage progenitor (GMP) stage of development.
Expression of C/EBP family members in freshly harvested C/EBPα-/- and C/EBPα+/+ and in HGF-treated FL cells
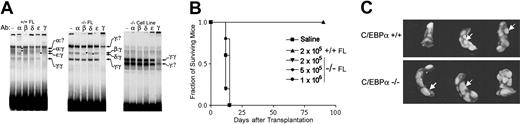
To determine whether other C/EBP proteins are expressed in C/EBPα+/+ and C/EBPα-/- primary FL cells and C/EBPα-/- FL cell lines, we examined nuclear lysates for the presence of C/EBPα, C/EBPβ, C/EBPδ, C/EBPϵ, and C/EBPγ by EMSA supershift assays. As shown in Figure 4A, DNA-protein complexes containing C/EBPα, C/EBPϵ, and C/EBPγ were detected in C/EBPα+/+ FL extracts. Two of the complexes appeared to correspond to heterodimers of C/EBPα or C/EBPϵ these proteins with C/EBPγ, an inhibitory C/EBP protein that preferentially dimerizes with other C/EBP family members in many cells.36-38 C/EBPα-/- FL contained neither C/EBPα nor C/EBPϵ but did have detectable levels of C/EBPβ and C/EBPδ binding species that occur as heterodimeric complexes with C/EBPγ. Examination of nuclear extracts from the SLF-IL-3-2 cell line (Figure 4A, right panel) revealed only C/EBPγ complexes, with little or no expression of the other 4 known C/EBP family members. Thus, C/EBPα and C/EBPγ are the major isoforms present in FL hematopoietic cells, and other C/EBPα activators do not appear to significantly compensate for the lack of C/EBPα in C/EBP-/- FL cells.
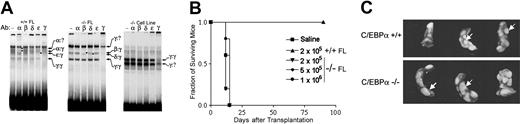
C/EBPα binding activities in FL extracts and C/EBPα-/- FL cell lines and transplantation of C/EBPα-/- FL cells in vivo. (A) Nuclear extracts prepared from wild-type FL (left panel), mutant FL (middle), or the C/EBPα-/- FL cell line cultured in IL-3 (right) were analyzed by EMSA using a consensus C/EBPα-binding site probe. Binding reactions included either nonimmune (NI) serum or the indicated C/EBPα antibodies. DNA-protein complexes were resolved on nondenaturing polyacrylamide gels. The identities of various dimeric C/EBPα species are shown by arrows; asterisks denote complexes that are supershifted by the antibody. (B) C/EBPα+/+ and C/EBPα-/- FL cells were transplanted into lethally irradiated mouse recipients at the indicated cell doses and monitored for survival according to the procedures described in “Materials and methods.” (C) In other groups, spleens were removed 10 to 12 days after transplantation and fixed in Tellesniczky solution to visualize spleen colonies (CFU-Ss) and photographed.
C/EBPα binding activities in FL extracts and C/EBPα-/- FL cell lines and transplantation of C/EBPα-/- FL cells in vivo. (A) Nuclear extracts prepared from wild-type FL (left panel), mutant FL (middle), or the C/EBPα-/- FL cell line cultured in IL-3 (right) were analyzed by EMSA using a consensus C/EBPα-binding site probe. Binding reactions included either nonimmune (NI) serum or the indicated C/EBPα antibodies. DNA-protein complexes were resolved on nondenaturing polyacrylamide gels. The identities of various dimeric C/EBPα species are shown by arrows; asterisks denote complexes that are supershifted by the antibody. (B) C/EBPα+/+ and C/EBPα-/- FL cells were transplanted into lethally irradiated mouse recipients at the indicated cell doses and monitored for survival according to the procedures described in “Materials and methods.” (C) In other groups, spleens were removed 10 to 12 days after transplantation and fixed in Tellesniczky solution to visualize spleen colonies (CFU-Ss) and photographed.
Hyperproliferation of C/EBPα-/- FL cells in vivo
To determine whether primitive hematopoietic progenitors in the C/EBPα-/- FL cell population show hyperproliferation in vivo, we transplanted C/EBPα-/- FL cells into lethally irradiated recipients at increasing cell doses and looked for the ability of these cells to give rise to macroscopic spleen colonies (CFU-Ss). As might be anticipated, radioprotection was not observed in animals receiving up to 1 × 106/cells C/EBPα-/- FL cells per mouse, because these cells are unable to give rise to mature neutrophils (Figure 4B). In comparison, C/EBPα+/+ FL cells were able to radioprotect all recipient mice and promote their short-term survival for 60 to 90 days with only 2 × 105 cells/mouse. Even though mice that received transplants with C/EBPα-/- FL cells do not survive lethal irradiation, there was a 4-fold increase in the number of progenitors that give rise to CFU-Ss in mice that received transplants with C/EBPα-/- FL compared with their littermate controls (Table 3). In addition, the size of the CFU-Ss derived from C/EBPα-/- transplanted FL cells were much larger than those CFU-Ss derived from the C/EBPα+/+ transplanted FL cells, indicating that the C/EBPα-/- CFU-Ss were hyperproliferating in vivo (Figure 4C). Thus, despite the fact that C/EBPα-/- FL cells give rise to increased numbers and size of CFU-Ss, they cannot confer radioprotection, presumably because of an inability to differentiate into functionally mature cells.
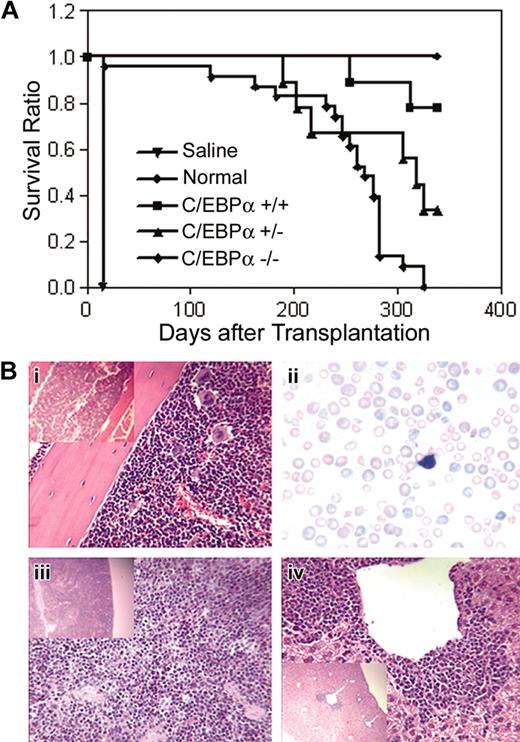
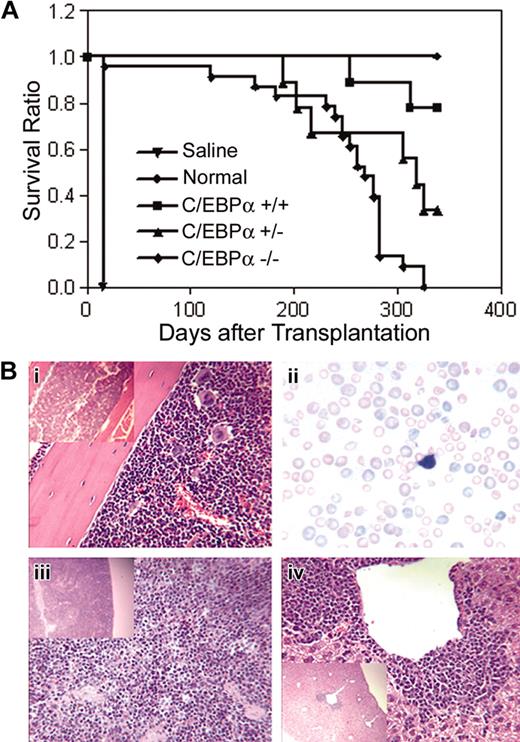
To determine whether C/EBPα-/- FL cells could engraft a compromised host and develop leukemia, C/EBPα-/-, C/EBPα+/-, and C/EBPα+/+ FL cells (Ly-5.1 donor) were transplanted into lethally irradiated mice together with normal BMCs (Ly-5.2 host cells) to provide short-term radioprotection. Groups of mice that received transplants were monitored over time for survival over a period of 339 days. The median survival of mice that received transplants with C/EBPα-/- FL cells was 268 days as compared with 339 days for the C/EBPα+/+ group and 318 days for the C/EBPα+/- group (Figure 5A). All C/EBPα-/- group mice were dead 325 days after transplantation, whereas 7 of 9 (77.8%) of C/EBPα+/+ group and 3 of 9 (33.3%) of the C/EBPα+/- group mice remained alive at the end of the experiment. Examination of PB smears of C/EBPα-/- mice showed marked anisocytosis and poikilocytosis of red cells, nucleated reds, frequent polychromasia, hyposegmented neutrophils, neutropenia, and immature leukocytes, indicating myelodysplastic syndrome (MDS) (Figure 5Bii). Histopathologic examination of moribund mice that received transplants with C/EBPα-/- FL cells revealed a variety of lesions, some of which were associated with opportunistic bacterial infections (skin or inner ear sores). In addition, BM hyperplasia with or without erythroid hyperplasia, increased blast cells, and paratrabecular clusters of megakaryocytes were detected in mice that received transplants with C/EBPα-/- FL cells (Figure 5Bi). In addition, splenic and hepatic extramedullary hematopoiesis was observed (Figure 5Biii,iv). Even though host BMCs were cotransplanted with C/EBPα-/- FL cells and the recipient mice showed mixed chimerism status, only the C/EBPα-/- mice had signs of MDS, suggesting that MDS might be the cause of increased mortality of the C/EBPα-/- group. These data also suggest C/EBPα-/- FL cells are hyperproliferative in vivo.
Survival of mice that received transplants with C/EBPα-/- FL cells and host-derived radioprotective cells. (A) C/EBPα+/+, C/EBPα+/-, and C/EBPα-/- FL cells were cotransplanted with host-derived radioprotective cells into lethally irradiated mice at cell doses described in “Materials and methods” and were monitored for survival. Photomicrographs of hematoxylin/eosin-stained sections from BM (i), spleen (iii), and liver (iv), and Giesema-stained peripheral blood smear (ii) of C/EBPα-/- mice are at magnifications of × 400 and insert × 100.
Survival of mice that received transplants with C/EBPα-/- FL cells and host-derived radioprotective cells. (A) C/EBPα+/+, C/EBPα+/-, and C/EBPα-/- FL cells were cotransplanted with host-derived radioprotective cells into lethally irradiated mice at cell doses described in “Materials and methods” and were monitored for survival. Photomicrographs of hematoxylin/eosin-stained sections from BM (i), spleen (iii), and liver (iv), and Giesema-stained peripheral blood smear (ii) of C/EBPα-/- mice are at magnifications of × 400 and insert × 100.
Defect in macrophage development in mice that received transplants of C/EBPα-/- FL cells
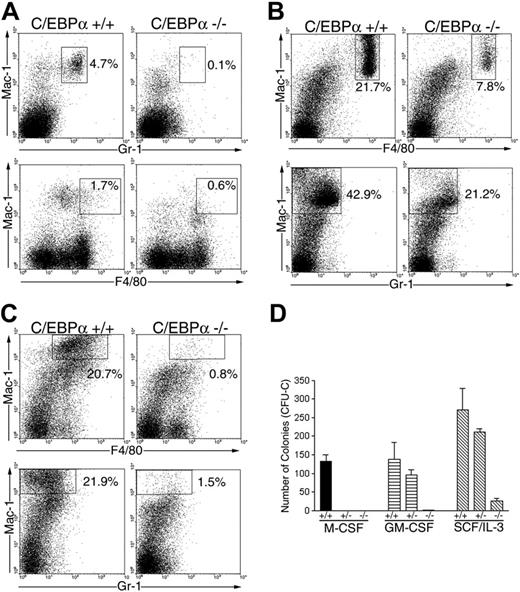
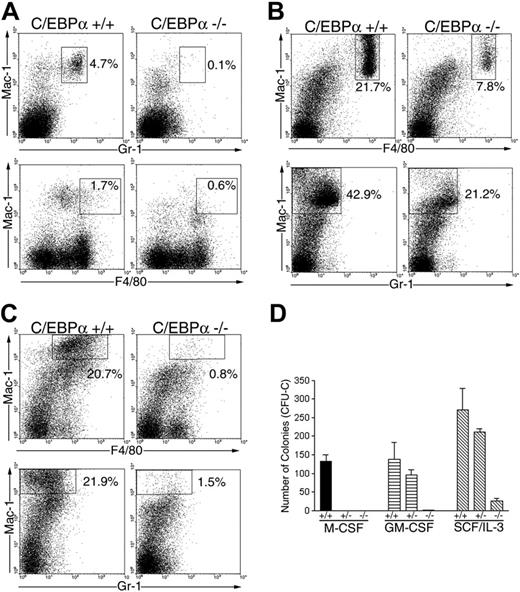
Because C/EBPα-/- FL could not differentiate into macrophages in response to M-CSF in vitro, we evaluated whether C/EBPα-/- FL could give rise to macrophages in vivo. In addition, we wanted to determine whether this developmental defect was intrinsic to the HPCs or their environment. Therefore, C/EBPα-/- and C/EBPα+/+ FL cells (Ly-5.1 donor) were transplanted into lethally irradiated mice together with normal BMCs (Ly-5.2 host cells) to provide radioprotection and track the development of donor-derived macrophages in vivo. First, we examined PB cells for the presence of donor-derived monocytes/macrophages and granulocytes by flow cytometry (Figure 6A). As expected,11 few if any donor-derived granulocytes (Gr-1Br Mac-1Br) (0.1%) were detected in PBCs of mice that received transplants with C/EBPα-/- FL cells, whereas 4.7% of the PBCs were Gr-1Br Mac-1Br granulocytes in mice that received transplants with C/EBPα+/+ FL cells. Also, mice that received transplants with C/EBPα-/- FL cells showed low levels (0.6%) of Mac-1br F4.80br macrophages compared with mice that received transplants with C/EBPα+/+ FL cells, which had 1.7% Mac-1br F4.80br macrophages.
Role of C/EBPα in macrophage differentiation in vivo. C/EBPα+/+ and C/EBPα-/- FL cells were transplanted into lethally irradiated mouse recipients in combination with normal BMCs. Four to 6 months after transplantation, peripheral blood (A), PECs (B), and thioglycollate-elicited PECs (C) were analyzed by 3-color flow cytometry for donor-derived (Ly-5.1) granulocyte and macrophage with use of Gr-1, F4/80, and Mac-1 according to the procedures outlined in “Materials and methods.” (D) BMCs were obtained from mice that received transplants, and FACS was performed for donor-derived cells and plated in colony assays in the presence of the indicated cytokines according to the procedures outlined in “Materials and Methods.” These data are presented as the mean colony formation ± the SE and are representative of 2 separate experiments.
Role of C/EBPα in macrophage differentiation in vivo. C/EBPα+/+ and C/EBPα-/- FL cells were transplanted into lethally irradiated mouse recipients in combination with normal BMCs. Four to 6 months after transplantation, peripheral blood (A), PECs (B), and thioglycollate-elicited PECs (C) were analyzed by 3-color flow cytometry for donor-derived (Ly-5.1) granulocyte and macrophage with use of Gr-1, F4/80, and Mac-1 according to the procedures outlined in “Materials and methods.” (D) BMCs were obtained from mice that received transplants, and FACS was performed for donor-derived cells and plated in colony assays in the presence of the indicated cytokines according to the procedures outlined in “Materials and Methods.” These data are presented as the mean colony formation ± the SE and are representative of 2 separate experiments.
To confirm the deficit in macrophages present in PB of mice that received transplants with C/EBPα-/- FL cells, we analyzed donor-derived macrophages in resident peritoneal exudates cells (PECs) and thioglycollate-elicited PEC populations. We found that 21.7% of the donor-derived resident PECs were Mac-1br F4.80br macrophages in mice that received transplants with C/EBPα+/+ FL cells, in contrast to 7.8% for C/EBPα-/- FL cells (Figure 6B). In addition, 20.7% of the donor-derived PEC population were Mac-1br F4.80br macrophages 3 days after thioglycollate treatment in mice that received transplants with C/EBPα+/+ FL cells compared with 0.8% for C/EBPα-/- FL cells (Figure 6C). Similarly, mice that received transplants with C/EBPα+/+ FL cells gave rise to more macrophages (21.9%) than mice that received transplants with C/EBPα-/- FL cells (1.5%) after thioglycollate treatment when Mac-1 and Gr-1 markers were assayed (Figure 6B-C).
To evaluate whether mice that received transplants with C/EBPα-/- FL cells lacked progenitors that proliferate in response to M-CSF, we obtained donor-derived (Ly-5.1) BMCs by FACS and plated them in soft agar colony assays in the presence of M-CSF, GM-CSF, and the combination of SCF and IL-3. Donor-derived BMCs from mice that received transplants with C/EBPα-/- FL cells lacked progenitors responsive to M-CSF in contrast to BMCs from mice that received transplants with C/EBPα+/+ FL cells, which showed significant colony formation in response to M-CSF (Figure 6D). Mice that received transplants with C/EBPα-/- FL cells also lacked progenitors responsive to GM-CSF and contained fewer colonies that responded to the combination of SCF/IL-3 in comparison to mice that received transplants with C/EBPα+/+ FL cells. Although we observed a decrease in the number of colonies that responded to SCF/IL-3 in C/EBPα-/- FL transplants, the remaining colonies in the plates were large hyperproliferative colonies similar to those observed when C/EBPα-/- FL was directly cultured in SCF/IL-3 (Figure 1). Collectively, these data show that C/EBPα-/- FL cells have an intrinsic defect in macrophage development in vivo.
Discussion
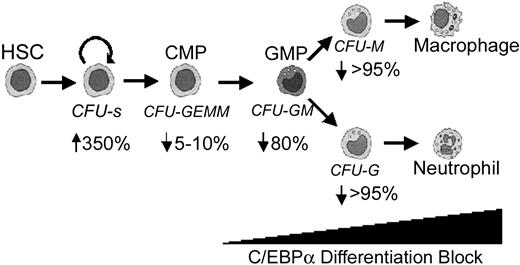
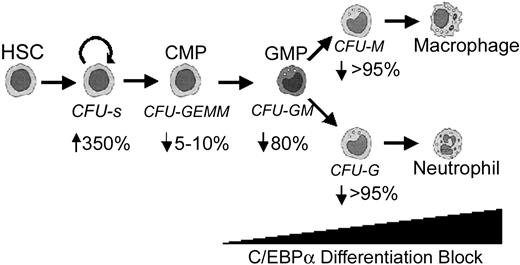
In this report we show that HPCs that lack C/EBPα are hyperproliferative, fail to differentiate, and show increased self-renewal potential in vitro. HGF-dependent progenitor cell lines can be directly derived from C/EBPα-/- FL cells in vitro, suggesting that C/EBPα may play a role in limiting the self-renewal of myeloid progenitor cells. In addition to the absence of G-CSF-responsive progenitors, we found that C/EBPα-/- FL lack M-CSF-responsive macrophage progenitors. This novel macrophage defect is intrinsic to the hematopoietic compartment because the development of donor-derived macrophage progenitors and their progeny is severely impaired in mice that received transplants with C/EBPα-/- FL. C/EBPα-/- FL cells hyperproliferate when transplanted in vivo and give rise to large spleen colonies (CFU-Ss), which correlates with the development of MDS. Finally, there is a significant loss in GM-CSF- and IL-3-responsive bipotential GM-progenitors (CFU-GM or GMP), whereas the multipotential colonies containing erythroid and megakaryocytes (CFU-GEMM [granulocyte, erythrocyte, megakaryocyte, macrophage colony-forming unit] or CMP) are unaffected. Therefore, we propose that C/EBPα is required for the CMP to differentiate into the GMP (Figure 7).
Summary of developmental defects in C/EBPα-deficient mice. Arrows indicate the percentage decrease in colony formation of C/EBPα-/- FL cells in vitro relative to control mice from data presented in Figure 1A. C/EBPα-/- FL cells contained more CFU-Ss than control mice did.
Summary of developmental defects in C/EBPα-deficient mice. Arrows indicate the percentage decrease in colony formation of C/EBPα-/- FL cells in vitro relative to control mice from data presented in Figure 1A. C/EBPα-/- FL cells contained more CFU-Ss than control mice did.
The defect in macrophage development in C/EBPα-/- FL progenitors was not observed in a previous study11 ; however, in that study, multiple HGFs were used to stimulate colony formation rather than M-CSF alone. It is possible that multiple HGFs may act differently in combination than M-CSF alone to drive macrophage proliferation and differentiation from more primitive progenitors, thereby bypassing the requirement for C/EBPα. Also, in the previous study, PB monocyte counts were used to determine the effect on the macrophage lineage in C/EBPα-/- mice compared with their wild-type littermates; however, we have found that accurate measurements of PB monocytes are difficult because of the low percentage of these cells in the PB (< 2%). Finally, our results suggest that defects in macrophage development might be due to a defect in M-CSFR expression, which was nearly absent in C/EBPα-/- FL cells. Although defects in M-CSFR were not observed in the previous study, C/EBPα in combination with PU.1 and AML1 has been shown to regulate M-CSFR expression.39,40 It is also possible that the absence of detectable G-CSFR and M-CSFR in C/EBPα-/- FL might reflect the severe reduction observed in the CFU-GM (GMP) population, thus resulting in the absence of M-CSF- and G-CSF-responsive progenitors and their differentiated progeny that would normally express M-CSFR and G-CSFR receptors.
It was unexpected that HGF-dependent hematopoietic cell lines could be directly derived by placing C/EBPα-/- FL in cultures containing HGF in vitro because alterations in oncogene and tumor suppressor gene expression pathways are normally required for immortalization of hematopoietic cells. Two previous studies showed that HPC lines could be established from C/EBPα-/- FL by infection with retroviruses carrying the Hox11 gene or the dominant-negative form of the retinoic acid receptor (dnRARα), but did not report immortalization without Hox11 or dnRARα.13,41 In both studies, the cell lines established were able to differentiate into neutrophils in response to GM-CSF plus IL-3, demonstrating that myeloid differentiation can occur by way of a C/EBPα-independent pathway. Although the expression of C/EBP family members was not analyzed during the differentiation of the dnRARα cell lines, GM-CSF/IL-3-induced differentiation of the Hox11 cell lines correlated with an increase in both C/EBPϵ and C/EBPβ expression, indicating that other C/EBP family members might substitute for C/EBPα. In contrast to the Hox11 and dnRARα cell lines, the SCF/IL-3 cell lines do not undergo further neutrophil differentiation in response to GM-CSF (data not shown). Because these cell lines do not express other C/EBP family members that might promote GM-CSF/IL-3-induced maturation, it is likely that these cell lines require C/EBPα to undergo further neutrophil differentiation. Because the cell lines developed in our study do not express additional immortalizing genes (eg, Hox11 or RARα), they may also represent better models to evaluate whether structurally modified C/EBPα proteins, or other C/EBP family members, can promote growth arrest and neutrophil differentiation. In addition, the cell lines provide a model for the identification of C/EBPα target genes required to drive these processes.
The results of the colony formation assay with C/EBPα-/- FL suggest that C/EBPα is required for the CMP to differentiate into the GMP (Figure 7). This is supported by the severe reduction in GM-CSF- and IL-3-responsive bipotential CFU-GM and more committed CFU-G and CFU-M progenitors. This maturation block is intrinsic to the hematopoietic cell compartment because BM progenitors derived from C/EBPα-/- FL cells are blocked at the same stage in differentiation when transplanted in vivo. Increased numbers of spleen colonies (CFU-Ss) were observed when C/EBPα-/- FL cells were transplanted in vivo, suggesting that C/EBPα may also act to limit the proliferation of more primitive progenitors. This observation is supported by the experiments showing enhanced secondary plating potential or increased self-renewal potential of C/EBPα-/- FL cells in vitro, which leads to the development of HGF-dependent cell lines blocked in differentiation. It is currently not known whether C/EBPα plays a role in limiting the proliferation of HSCs and whether there are increased numbers of HSCs in C/EBPα-/- FL. However, this question can be addressed by using the competitive repopulation assay with C/EBPα-/- FL and BMCs to more precisely determine the number of stem cells present.
C/EBPα-/- FL cells showed a hyperproliferative response to signals in vivo as demonstrated by the proliferation of large CFU-Ss and the MDS phenotype. Therefore, we hypothesize that transplantation of C/EBPα-/- FL might give rise to leukemia over time, although the acquisition of additional mutations may be required for leukemogenesis.
Future experiments are planned to examine whether MDS or leukemia can be accelerated in mice that receive transplants by increasing the number of C/EBPα-/- FL cells with decreased numbers of radioprotective BMCs. Alternatively, to avoid a potential host immune response during leukemia development we will transplant C/EBPα-/- FL cells into immune compromised animals. To begin to evaluate what additional mutations might be critical for leukemogenesis in this model, we have determined in preliminary experiments that HGF-dependent cell lines can be directly established from PBCs and BMCs of mice that received transplants with C/EBPα-/- FL cells but not C/EBPα+/+ FL cells (data not shown). Thus, it will be possible to establish HGF-dependent cell lines from mice that received transplants with C/EBPα-/- over time and compare gene expression patterns between cell lines by microarray or other analysis to identify additional mutations.
In summary, these observations support the hypothesis that C/EBPα plays a critical role in regulating the balance between proliferation and growth arrest/differentiation in hematopoietic progenitors. Furthermore, transplantation of C/EBPα-/- FL cells into irradiated recipients supports the hypothesis that loss of C/EBPα expression or function may contribute to the differentiation block, enhanced proliferation, and development of some AML.25 This model may provide a means to determine what additional genetic events are required for loss of growth control during leukemogenesis.
Prepublished online as Blood First Edition Paper, April 8, 2004; DOI 10.1182/blood-2003-11-3963.
Supported in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (contract no. NO1-CO-12400).
The publisher or recipient acknowledges the right of the U.S. Government to retain a nonexclusive, royalty-free license in and to any copyright covering this article.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Barbara Shankle, Mehrnoosh Abshari, Kathleen Noer, Roberta Matthai, and Steve Stull for the outstanding technical assistance. We also thank Drs Simon Williams, Rich Schwartz, and Joost Oppenheim for their critical review of this manuscript.

![Figure 1. Effect of hematopoietic growth factors (HGFs) on the growth of C/EBPα-/- fetal liver (FL) cells in vitro. (A) FL cells were obtained from E16-E17 C/EBPα-/- mice and seeded into soft agar colony assays at 5 × 104 cells/mL in the presence or absence of 50 ng/mL human G-CSF (Amgen, Thousand Oaks, CA), 100 ng/mL M-CSF, 100 ng/mL murine SCF, 50 ng/mL murine GM-CSF, or 30 ng/mL murine IL-3 (Peprotech, Rocky Hill, NJ). (B) Bright field photomicrographs (× 16) of the 35-mm gridded (2 mm × 2 mm) plates were taken after 7 days. Dishes were incubated in a fully humidified atmosphere at 37°C, 5% CO2, and then scored for colony formation after 7 to 10 days (culture colony-forming unit [CFU-C]). The results are presented as the mean number of colonies plus or minus the standard error, and are representative of at least 3 separate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/6/10.1182_blood-2003-11-3963/6/m_zh80180466530001.jpeg?Expires=1767955585&Signature=fYUtZ7HAsOUmHi5BPNBpAMvi-qi7Tal38UOwhHQyPAq-OTmPs4zFa8BsnXGb2eS2GSNnTqm0RyCWut4rclwOakiQkZgJPsew6jmnmWfLjGAwrXM4OVYYSrbT6qRHqULrWzgrojoXho3yocjJD5hWCf9BAOgih7m6UOKSweWDs2Z9vaAAbcVYtH5twa26fTC80FD6TcnGLiYcpSlsqAOTHMX5Oof85Ff44bM7zpk~AIjgPj0FyFkE6eLZzn8tSV7k3~b0FXLw5FIYJnf5am~eHFUQ5jfuroMPwox6aOOay-HVE8S~53m6jNUoKwi0HFJoncxeC-hUdeIotgErHm44Og__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. Effect of hematopoietic growth factors (HGFs) on the growth of C/EBPα-/- fetal liver (FL) cells in vitro. (A) FL cells were obtained from E16-E17 C/EBPα-/- mice and seeded into soft agar colony assays at 5 × 104 cells/mL in the presence or absence of 50 ng/mL human G-CSF (Amgen, Thousand Oaks, CA), 100 ng/mL M-CSF, 100 ng/mL murine SCF, 50 ng/mL murine GM-CSF, or 30 ng/mL murine IL-3 (Peprotech, Rocky Hill, NJ). (B) Bright field photomicrographs (× 16) of the 35-mm gridded (2 mm × 2 mm) plates were taken after 7 days. Dishes were incubated in a fully humidified atmosphere at 37°C, 5% CO2, and then scored for colony formation after 7 to 10 days (culture colony-forming unit [CFU-C]). The results are presented as the mean number of colonies plus or minus the standard error, and are representative of at least 3 separate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/6/10.1182_blood-2003-11-3963/6/m_zh80180466530001.jpeg?Expires=1767955586&Signature=scrmOoNUel9U-xJT5sGMWNaKvRbqJavULLKWA~HiMC0KqlxYSicJVSnISFuBAJUq-eW0eu24WmAQIU95Le35zCBPQTKEEc8VI1dloBzeKjFsqcyOZN9nq6S6vWexIhwvfLRHF-t8mox7D-Se3C1MKLCz3pEyKOzsj2uRQhgKhur018~tIMJgBjGQ1bdrY2uXkSJAE9gmVOBuDVYN1wBIcr8Nl394zIUEwIT-M2rdYngtvIUwvywrAa27Lt7Zf1FTTkDUDHTekYfB9FcCrtGNaDxGS37Ss5xIvMeIccubgafUzw1jDwoOBU3ciPZJL11Wu9ZMVji2KVwo~yDEJiWIMQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)