Abstract
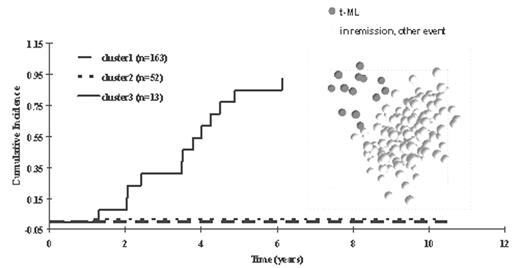
In children with acute lymphoblastic leukemia (ALL), failure due to therapy-related myeloid leukemia (t-ML) is a devastating complication. Using a target gene approach, only a few host genetic risk factors for t-ML have been defined. Microarray analysis of gene expression allows for a more genome-wide approach to identify possible genetic risk factors for t-ML. We assessed gene expression profiles (12625 gene probe sets) using oligonucleotide-based arrays in diagnostic ALL blasts from 228 children treated on St. Jude ALL protocols (Total XIII) that included etoposide; 13 of these children developed t-ML. A group of 83 probe sets were significantly related to the time-dependent risk of t-ML, with principal component analysis plot (right panel) separating patients who developed t-ML from the others. Hierarchical clustering of the 83 probe sets grouped patients into 3 clusters (n=163, n=52, n=13), with the cumulative incidence of t-ML being significantly higher in the last cluster (p < 0.0001, left panel) compared to those of the other gene-expression-defined clusters.
A permutation test indicated that probe sets selected by chance are unlikely to obtain the observed distinct clusters (p=0.045). Distinguishing genes included transcription-related oncogenes (v-Myb, Pax-5), cyclins (CCNG1, CCNG2 and CCND1) and Histone H4. Common transcription factor recognition elements among similarly up- or down-regulated genes included several involved in hematopoietic differentiation or leukemogenesis (Maz, PU.1, FOXO4). This approach has identified several genes whose expression differentiates patients at risk of t-ML, and provides targets for assessing the germline predisposition to leukemogenesis.
Author notes
Corresponding author