Abstract
In Epstein-Barr-virus (EBV)–positive lymphomas in immunocompetent patients, release of EBV DNA from tumor cells into the plasma might be useful for disease monitoring and prognostication. To test this hypothesis, we quantified serially plasma EBV DNA by quantitative polymerase chain reaction in 39 cases of EBV-positive (natural killer [NK] cell, n = 23; T cell, n = 8; B cell, n = 4; Hodgkin, n = 4) lymphomas. As control, EBV DNA was undetectable in 34 cases of EBV-negative lymphomas at diagnosis and during chemotherapy. In all cases of EBV-positive lymphomas, EBV DNA was detectable (105-1010 copies/mL) at diagnosis. It paralleled the clinical course, with EBV DNA becoming undetectable at remission and remaining elevated in refractory disease. On multivariate analysis, high-presentation EBV DNA (> 7.3 × 107 copies/mL) was significantly associated with an inferior overall survival (OS). Subgroup analysis of NK cell lymphomas, the largest cohort in this study, showed that presentation EBV DNA was correlated with disease stage and lactate dehydrogenase. On multivariate analysis, high-presentation EBV DNA (> 6.1 × 107 copies/mL) was significantly associated with an inferior disease-free survival. During treatment, patients with EBV DNA that showed further increases or failed to become undetectable had significantly inferior OS. In EBV-positive lymphomas, plasma EBV DNA is valuable as a tumor biomarker and for prognostication.
Introduction
Epstein-Barr virus (EBV) is a ubiquitous herpes virus that infects preferentially B cells and occasionally other cell types, especially epithelial cells.1 After primary infection, EBV establishes an asymptomatic latency state. Rarely, EBV infection may be associated with the development of malignancies.2 Hematologic malignancies associated with EBV infection include Burkitt lymphoma, Hodgkin lymphoma, some cases of Band T-cell lymphomas,3 natural killer/T (NK/T) cell lymphoma,4 primary effusion lymphoma,5 and pyothorax-associated lymphoma.6 Epithelial tumors related to EBV infection include nasopharyngeal carcinoma7 and gastric cancer.8
In immunocompromised patients, EBV infection is associated with another spectrum of malignancies. In organ allograft recipients receiving immunosuppression to prevent graft rejection or graft-versus-host disease, reactivation of EBV infection may lead to posttransplantation lymphoproliferative disorders (PTLDs).9-11 In patients with the acquired immunodeficiency syndrome who develop lymphomas, EBV infection is found in about 30% of centroblastic and up to 90% of immunoblastic lymphomas.12
In addition to its putative tumorigenic role, EBV has also been a target for disease monitoring. After organ allografting, increasing loads of EBV in whole blood, lymphocytes, and plasma are associated with corresponding increases in the risk of PTLD.13-17 In these cases, the lymphoproliferative disease is due to an expansion of cells directly transformed by EBV and growing opportunistically in the absence of an effective T-cell control.9 Therefore, the increase in circulating EBV DNA has been attributed to EBV replication.
In another EBV-associated malignancy, nasopharyngeal carcinoma (NPC), increases in circulating EBV have also been correlated with tumor load and disease activity.18,19 However, these patients are not immunocompromised, so that the increase in EBV DNA is unrelated to EBV reactivation. Rather, it has been proposed that fragments of the EBV genome are released from tumor cells into the circulation,18 so that quantification of EBV DNA might be a surrogate biomarker of tumor load. Plasma EBV DNA at presentation of NPC has been found to be an important prognostic marker.19
In lymphomas, the quantification of EBV DNA has so far been applied to the follow-up of immunocompromised patients with PTLD, where EBV reactivation occurs.13-17 In this study, we surmised that in EBV-related lymphomas in immunocompetent patients, an increase in circulating EBV DNA might also occur, owing to the release of EBV DNA from lymphoma cells. To test this hypothesis, plasma EBV DNA was quantified by a real-time quantitative polymerase chain reaction (Q-PCR) in a cohort of patients with EBV-related lymphoid malignancies at diagnosis and during follow-up. To ensure that the quantification of EBV during treatment might represent tumor load, it would be important to exclude a possible reactivation of EBV owing to the immunosuppressive effects of chemotherapy. To this end, patients with EBV-negative lymphomas receiving chemotherapy were tested as a model of mild immunosuppression, and patients undergoing hematopoietic stem cell transplantation (HSCT) were tested as a model of intense immunosuppression.
Patients, materials, and methods
Patients and sample preparation
Consecutive lymphoma patients diagnosed during a 6-year period (1998-2003) were prospectively studied. Lymphomas were classified according to the World Health Organization classification criteria.20 The presence of EBV was confirmed by in situ hybridization for EBV-encoded RNA (EBER) in the neoplastic cells on histopathologic examination. Cell-free plasma was separated immediately after venipuncture by centrifugation at 1200g for 15 minutes, and frozen at –20° C until assay. DNA was extracted from 500 μL plasma in a QIAamp DNA mini blood kit (Qiagen GmbH, Hilden, Germany) and eluted with 100 μL AE buffer (Qiagen). For each experiment, 5 μL DNA eluate (or 25 μL plasma volume equivalent) was used. The protocol was approved by the institutional review board at Queen Mary Hospital, Hong Kong.
Quantitative polymerase chain reaction (Q-PCR)
Q-PCR was performed by a real-time PCR assay with the ABI Prism 7700 Sequence Detector (PE Biosystems, Foster City, CA). PCR primers for the EBV gene EBNA1 (forward primer Ef, 5′-TCA TCA TCA TCC GGG TCT CC-3′, and reverse primer Er, 5′-CCT ACA GGG TGG AAA AAT GGC-3′) and the TaqMan probe (Ep, 5′-CGC AGG CCC CCT CCA GGT AGAA-3′) dual-labeled at the 5′ end with 6-carboxyfluorsecein (FAM) and the 3′ end with 6-carboxytetramethylrhodamine (TAMRA) were designed by the Primer Express software (PE Biosystems). The amplicon size of the EBNA1 Q-PCR product was 68 base pair (bp), being optimized by the Primer Express software. Q-PCR was set up in a volume of 50 μL with use of the TaqMan Universal PCR Master Mix (PE Biosystems), containing 5 μL purified DNA, 300 nM Ef/Er, and 200 nM Ep. Thermal cycling was initiated with a 2-minute incubation at 50° C, followed by a first denaturation step of 10 minutes at 95° C, and then 40 cycles of 95° C for 15 seconds (denaturation) and 60° C for 1 minute (reannealing and extension). Real-time PCR data were collected continuously and analyzed with the Sequence Detection System (PE Biosystems). Cycle thresholds (CT), at which a significant increase in fluorescence signal was first detected, were set at a minimum of 10 standard deviations above the mean baseline fluorescence, calculated from cycles 1 to 15. As the larger the starting quantity of target sequence there was, the earlier a significant increase in fluorescence would be observed, the CT was inversely proportional to the starting target gene copy number.
Cloning of plasmid pB-EBNA1 containing EBNA1 gene sequences
A fragment of the EBV EBNA1 gene detected by Q-PCR was amplified from an EBV-immortalized lymphoblastoid cell line with appropriate primers (5′-(CAU)4 AGG CCC AGG AGT CCC AGT A-3′ and 5′-(CUA)4 CTG CTC TAT CGC TCC CGG-3′). The PCR products were cloned into the pAMP1 vector (Gibco BRL, Life Technologies, Rockville, MD) to give the plasmid pB-EBNA1. The sequence of the cloned EBNA1 gene fragment was confirmed by DNA sequencing in both directions (ABI Prism 377; PE Biosystems).
Quantification of EBV EBNA1 gene
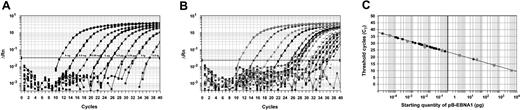
Standard curves were constructed by plotting the CTs against the logarithm of the starting amount of a serial dilution of pB-EBNA1, over an 8-log range from 5 ng to 0.5 fg (Figure 1A). Patient samples were tested in triplicates. The amplification profiles of the patient samples were similar to that of the plasmid standards (Figure 1A-B). The CTs of the patient samples were determined, and the initial starting sequence amount was calculated from the standard curve (Figure 1C). EBV genome equivalent copies were calculated with the following formula21 : copy number = X × 10–12 × (660 NP)–1 × 6.023 × 1023, where X is the amount in picograms, 660 is the average molecular weight of a nucleotide pair, NP is the number of nucleotide pairs in the plasmid (NP = 4289 for pB-EBNA1), and 6.023 × 1023 is the number of gene copies in one mole of plasmid.
Standard curves for quantification of EBV DNA. (A) Amplification plot for reactions with known starting amounts of pB-EBNA1 (an 8-log dilution from 5 ng to 0.5 fg, denoted by boxes next to the corresponding curves). Cycle number was plotted against change in normalized reporter signal (ΔRn). For each reaction, the fluorescence signal of the reporter dye (FAM) was divided by the fluorescence signal of the passive reference dye (ROX) to obtain a ratio defined as the normalized reporter signal (Rn). (B) Amplification plot of patients' plasma samples, demonstrating similar amplification profiles as the control plasmid shown in panel A. (C) Plot of standard curve of starting pB-EBNA1 amount against CT. Gray circles represent pB-EBNA1 standards as shown in panel A. Black circles represent patient samples. A standard curve was constructed for each assay. Note that as most samples showed moderate amounts of EBV DNA, patient samples tended to cluster on the left half of the graph. However, one patient with a high EBV DNA load is shown on the right half of the graph.
Standard curves for quantification of EBV DNA. (A) Amplification plot for reactions with known starting amounts of pB-EBNA1 (an 8-log dilution from 5 ng to 0.5 fg, denoted by boxes next to the corresponding curves). Cycle number was plotted against change in normalized reporter signal (ΔRn). For each reaction, the fluorescence signal of the reporter dye (FAM) was divided by the fluorescence signal of the passive reference dye (ROX) to obtain a ratio defined as the normalized reporter signal (Rn). (B) Amplification plot of patients' plasma samples, demonstrating similar amplification profiles as the control plasmid shown in panel A. (C) Plot of standard curve of starting pB-EBNA1 amount against CT. Gray circles represent pB-EBNA1 standards as shown in panel A. Black circles represent patient samples. A standard curve was constructed for each assay. Note that as most samples showed moderate amounts of EBV DNA, patient samples tended to cluster on the left half of the graph. However, one patient with a high EBV DNA load is shown on the right half of the graph.
PCR precautions
All precautions preventing contamination were rigorously adhered to.21 DNA extraction, cloning of pB-EBNA1, and Q-PCR were performed in 3 different laboratories. All samples were handled with positive displacement pipettes with aerosol-resistant tips. Negative blanks included in each PCR did not give positive results.
Quality assurance of Q-PCR
To control for plate-to-plate variation in PCR efficiencies, so that all results were comparable, a standard curve was constructed in each Q-PCR experiment with serial dilutions of the same stock solution of pB-EBNA1, which was prepared at the start of the project and frozen at –20° C in small aliquots for single use only.
Statistical analysis
Overall survival (OS) was measured from diagnosis to death or last follow-up. Disease-free survival (DFS) was measured from complete remission (CR) to the date of relapse. The International Prognostic Index (IPI) was calculated according to published criteria.22 To evaluate prognostic significance, the IPI, age (older or younger than 60 years), sex, lactate dehydrogenase (LDH) level (above or below 2 times the upper limit of normal, 550 IU/L), stage (I/II and III/IV), and diagnostic EBV DNA level (above or below the median EBV DNA level) were analyzed with a Kaplan Meier model using logistic regression computation (SPSS 7.0; SPSS, Chicago IL).
Results
Patients and controls
A total of 39 patients with EBV-positive lymphomas were investigated (Table 1), together with 7 patients with PTLD. Thirty-four cases of EBV-negative lymphomas, 16 patients after allogeneic HSCT (donors, 7 human leukocyte antigen [HLA]–matched unrelated, 4 HLA-identical sibling, 2 HLA-matched parents) were also studied. A total of 427 patient plasma samples were assayed with Q-PCR, of which 378 were from patients with EBV-positive lymphoma and from 49 patients with EBV-negative lymphoma.
Increases of EBV DNA during immunosuppression
In all the 34 cases of EBV-negative lymphomas, EBV DNA was undetectable at diagnosis, during chemotherapy, or at any stage of the illness (Table 2). However, in patients after HSCT, EBV DNA was detectable in 10 of 16 cases, which ranged from 8.5 × 105 to 1.6 × 108 copies/mL (median, 5.8 × 106 copies/mL). This had all occurred during immunosuppression (antithymocyte globulin, n = 4; high-dose intravenous steroid, n = 10) for graft-versus-host-disease (GVHD) (grade II, n = 2; grade III, n = 4; grade IV, n = 4). However, none of these patients subsequently developed any EBV-related lymphoproliferative disorders. In patients who recovered from GVHD, EBV DNA eventually declined to undetectable levels after cessation of immunosuppression. However, in patients who died of GVHD, EBV DNA remained elevated until death. Therefore, EBV DNA was not increased during the mild immunosuppression typically associated with conventional chemotherapy but might be elevated during intense immunosuppression.
Posttransplantation lymphoproliferative disorder
Seven cases of PTLD after organ allografting (bone marrow, n = 3; kidney, n = 3; liver, n = 1) were studied (Table 2). The median EBV DNA at presentation was 3.1 × 107 copies/mL. PTLD after HSCT did not differ from PTLD after solid organ allografting (median, 5.1 × 109 versus 0.9 × 108 copies/mL, P = .16). For cases that responded to treatment (n = 3), EBV DNA fell from a median peak of 3.4 × 107 copies/mL to undetectable when CR was reached. However, for refractory cases (n = 4), EBV DNA remained elevated, ranging from 9.1 × 108 to 1.4 × 1011 copies/mL until death.
NK cell lymphoma
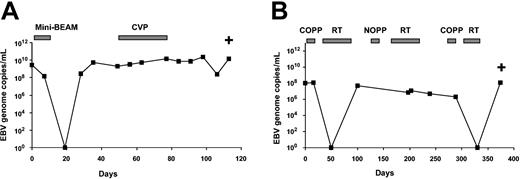
Twenty-three cases of NK cell lymphomas were studied. The median EBV DNA at presentation was 6.1 × 107 copies/mL (range, 2.7 × 105 to 7.1 × 1010). NK cell lymphomas might be classified clinically into nasal, nonnasal, and disseminated subtypes. These subtypes, however, were associated with similar median EBV DNA at presentation (Table 3). Two patients died shortly after diagnosis. For 21 patients serially monitored, EBV DNA paralleled treatment responses. In 7 cases that achieved CR, EBV DNA became undetectable. However, in 14 refractory or relapsing cases, EBV DNA remained high (range, 6.7 × 107-2.4 × 1011; median, 2.8 × 108 copies/mL) with persistent disease and until death (Figure 2A-B). Clinicopathologic correlations showed that plasma EBV DNA at presentation was positively correlated with disease stage and LDH (Table 3). The peak EBV DNA level, defined as the highest level in the clinical course of the patients (in 10 cases this was the presentation level, and in 13 cases a further peaking of EBV DNA occurred during treatment) was also positively correlated with advanced stage (Table 3). As patients in stage III/IV have much more widespread tissue involvement than patients in stage I/II, the 20- to 30-fold increase in EBV DNA in the former as compared with the latter group most likely reflected a much larger tumor load.
Changes of plasma EBV DNA during treatment of NK cell lymphomas. (A) A case of nasal NK cell lymphoma. An initial response to chemotherapy was followed by a rapid drop of EBV DNA to undetectable levels. However, with disease relapse, there was a resurgence of EBV DNA. The patient did not respond to treatment, and EBV DNA remained elevated until death. (B) A case of nonnasal NK cell lymphoma. Initial response to treatment was followed by a fall in EBV DNA. With relapse, there was increase in EBV DNA. Although there was a brief response with undetectable EBV DNA, a relapse associated with elevation of EBV DNA occurred, finally leading to death. Mini-BEAM indicates carmustine, etoposide, cytosine arabinoside, melphalan; CVP, cyclophosphamide, vincristine, prednisolone; COPP, cyclophosphamide, vincristine, procarbazine, prednisolone; RT, radiotherapy; NOPP, mitoxantrone, vincristine, procarbazine, prednisolone; and +, death.
Changes of plasma EBV DNA during treatment of NK cell lymphomas. (A) A case of nasal NK cell lymphoma. An initial response to chemotherapy was followed by a rapid drop of EBV DNA to undetectable levels. However, with disease relapse, there was a resurgence of EBV DNA. The patient did not respond to treatment, and EBV DNA remained elevated until death. (B) A case of nonnasal NK cell lymphoma. Initial response to treatment was followed by a fall in EBV DNA. With relapse, there was increase in EBV DNA. Although there was a brief response with undetectable EBV DNA, a relapse associated with elevation of EBV DNA occurred, finally leading to death. Mini-BEAM indicates carmustine, etoposide, cytosine arabinoside, melphalan; CVP, cyclophosphamide, vincristine, prednisolone; COPP, cyclophosphamide, vincristine, procarbazine, prednisolone; RT, radiotherapy; NOPP, mitoxantrone, vincristine, procarbazine, prednisolone; and +, death.
EBV-positive non-Hodgkin lymphoma (NHL)
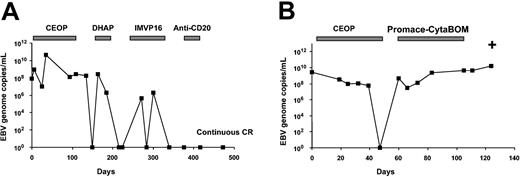
Twelve EBV-positive NHLs were studied (Table 2). The median EBV DNA at presentation, as well as the peak EBV DNA, was comparable in angioimmunoblastic T-cell lymphoma, diffuse large B-cell lymphoma, and peripheral T-cell lymphoma. Similar to NK cell lymphomas, the EBV DNA changed with treatment in both B- and T-cell lymphomas (Figure 3A-B), with 5 cases reaching CR having undetectable EBV DNA, and 7 refractory cases maintaining a high EBV DNA (2.3 × 106-1.5 × 1010 copies/mL) until death.
Changes of plasma EBV DNA during treatment of EBV-positive lymphomas. (A) A case of diffuse large B-cell lymphoma. EBV DNAdropped to undetectable levels after initial chemotherapy, However, with relapses, EBV DNA rose again to significant levels. Successful treatment with salvage chemotherapy led to persistently undetectable levels of EBV. The patient had remained in continuous complete remission. (B) A case of angioimmunoblastic T-cell lymphoma. EBV dropped to undetectable levels after initial chemotherapy. On relapse, there was no response to chemotherapy and EBV DNA remained elevated until death. CEOP indicates cyclophosphamide, epirubicin, vincristine, prednisolone; DHAP, cisplatin, cytosine arabinoside, dexamethasone; IMVP16, ifosfamide, methotrexate, etoposide; anti-CD20, rituximab, ProMACE-CytaBOM: cyclophosphamide, adriamycin, etoposide, prednisolone, cytosine arabinoside, bleomycin, vincristine, methotrexate; and +, death.
Changes of plasma EBV DNA during treatment of EBV-positive lymphomas. (A) A case of diffuse large B-cell lymphoma. EBV DNAdropped to undetectable levels after initial chemotherapy, However, with relapses, EBV DNA rose again to significant levels. Successful treatment with salvage chemotherapy led to persistently undetectable levels of EBV. The patient had remained in continuous complete remission. (B) A case of angioimmunoblastic T-cell lymphoma. EBV dropped to undetectable levels after initial chemotherapy. On relapse, there was no response to chemotherapy and EBV DNA remained elevated until death. CEOP indicates cyclophosphamide, epirubicin, vincristine, prednisolone; DHAP, cisplatin, cytosine arabinoside, dexamethasone; IMVP16, ifosfamide, methotrexate, etoposide; anti-CD20, rituximab, ProMACE-CytaBOM: cyclophosphamide, adriamycin, etoposide, prednisolone, cytosine arabinoside, bleomycin, vincristine, methotrexate; and +, death.
Hodgkin lymphoma
Seventeen consecutive cases of Hodgkin lymphoma (nodular sclerosing, n = 15; mixed cellularity, n = 1, lymphocyte depleted, n = 1) were studied, of which 4 cases (all nodular sclerosing) were EBER positive, and 13 cases were EBER negative. EBV DNA was detectable in all the EBER-positive cases and undetectable in any of the EBER-negative cases (Table 2). For the 4 EBER-positive cases, EBV DNA became undetectable in 3 patients who achieved CR and remained negative at the latest follow-up. In the last patient, death because of sepsis occurred shortly after the initiation of chemotherapy, so that changes in EBV DNA could not be evaluated.
Prognostic significance of EBV DNA for the whole group of EBV-positive lymphoma
This series comprised lymphomas of various histologic subtypes, so that the prognosis and treatment were heterogeneous. Excluding PTLD, the median presentation EBV DNA of all 39 cases of EBV-positive lymphoma was 7.3 × 107 copies/mL. This median presentation EBV DNA level was found to be a significant cutoff. Patients with EBV DNA of more than 7.3 × 107 copies/mL (n = 20), when compared with those with EBV DNA of less than or equal to 7.3 × 107 copies/mL (n = 19), had significantly inferior OS (2.0 versus 30.6 months, P = .0015) and DFS (0.5 versus 8.7 months, P = .0023). When presentation EBV DNA (≤ versus > 7.3 × 107 copies/mL), disease stage (I/II versus III/IV), age (≤ versus > 60 years), and LDH (≤ versus > 2 times normal) were entered into multivariate analysis, EBV DNA remained the only significant factor negatively affecting OS (P = .041). For DFS, only advanced-stage (P = .067) and high EBV DNA (P = .066) showed a trend toward significance.
Prognostic significance of EBV DNA in NK cell lymphoma
Subgroup analysis was performed for NK cell lymphoma, the most prevalent EBV-positive lymphoma in the Chinese population, and the largest group in this series (Table 4). The prognostic effect of the clinicopathologic factors at presentation, including clinical subtypes, age, stage, LDH, IPI, and EBV DNA, was evaluated (Table 4). On univariate analysis, the DFS was significantly related to the clinical subtypes, stage, and the presentation EBV DNA (with the median presentation level of all the cases of NK cell lymphomas, 6.1 × 107 copies/mL, as a cutoff). On multivariate analysis, only the presentation EBV DNA remained as a significant factor negatively affecting DFS (P = .025). Similarly, the OS was significantly related to clinical subtypes, stage, and presentation EBV DNA on univariate analysis. On multivariate analysis, only the presentation EBV DNA showed a trend toward negatively affecting OS (P = .06). As EBV DNA changed during treatment, the prognostic significance of 2 dynamic parameters was also evaluated (Table 5). Two patients died soon after diagnosis, so that only 21 patients could be serially assessed. Patients who showed a rise of EBV DNA that exceeded the presentation level at any stage of their disease (n = 13) had a significantly inferior median OS as compared with those who did not have a rise (n = 8). Furthermore, patients whose EBV DNA never became undetectable at any stage of their illness (n = 5, all dead from disease) had a significantly inferior median OS as compared with those who had an undetectable EBV DNA during some stage in their clinical course (n = 16, 7 alive and in continuous remission).
Discussion
This is to date the first comprehensive study of plasma EBV DNA as a surrogate tumor biomarker in a wide variety of EBV-related lymphoid malignancies in immunocompetent patients. Previous studies have only focused on the quantification of EBV in EBV-related PTLD after organ allografting. In these immunocompromised patients, reactivation of latent EBV infection was considered an important pathogenetic mechanism. In fact, reactivation of EBV infection was found to be frequent in patients after allogeneic HSCT. Patients with low levels of EBV might remain asymptomatic. However, those with high EBV load had a much-increased risk of PTLD.16,17 In these cases, the monitoring of plasma EBV DNA might be a direct measurement of viral activity. Other studies have, however, measured the number of EBV-infected lymphocytes,13,22 or EBV-infected tumor cells,14 which reflected viral reactivation or the direct presence of circulating lymphoma cells. Both approaches are valid in the setting of organ allografting, as a lack of immune control of EBV proliferation is the underlying mechanism.
In patients with EBV-related lymphomas who are immunocompetent, an unchecked EBV proliferation is unlikely to happen. Indeed, in EBV-related solid tumors, circulating EBV DNA exists as short fragments of less than 200 bp.23 This finding implies that the increase of EBV DNA is due to tumor release of EBV fragments instead of virion reactivation. Similarly, the circulating EBV DNA in patients with lymphoma has also been shown to be about 180 bp.23 We, therefore, hypothesized that the release of EBV from EBV-positive lymphoma cells might be a general phenomenon and could be a target for disease monitoring. This proposition was validated by the finding that plasma EBV DNA was detectable at high levels in all the EBV-positive lymphomas at diagnosis. None of the EBV-negative lymphomas showed demonstrable plasma EBV DNA. Interestingly, the presenting levels of EBV DNA in EBV-positive lymphomas (medians ranging from 106 to 109 copies/mL) were comparable with those in PTLD after allografting (median at 107 copies/mL). Therefore, plasma EBV DNA, whether derived putatively from the lymphoma or from reactivation of EBV infection, appears to be significantly elevated at diagnosis, thereby providing a useful biomarker for EBV-related lymphoid malignancies.
To further confirm this hypothesis, the plasma EBV DNA levels were tested serially through therapy. We first showed that the mild degree of immunosuppression found in chemotherapy was not associated with an increase in EBV DNA, validating that during treatment any detectable EBV DNA was tumor derived and not due to reactivation of latent EBV infection. Furthermore, we showed that EBV DNA levels paralleled the clinical course of the patients. For patients responding to chemotherapy, there was a rapid decline of EBV DNA, sometimes from very high levels to undetectable within a few weeks. As plasma DNA has a rapid clearance, with a half-life only in minutes,24,25 the measurement of EBV DNA may in fact give a much quicker and convenient measurement of tumor mass, as compared with more conventional serologic measurements or imaging studies. These observations were comparable with those in PTLD. Lucas et al14 showed that successful treatment of PTLD was associated with corresponding falls in EBV DNA. Similarly, Stevens et al,15 using unfractionated blood, showed that EBV DNA correlated with disease activity in patients with multiple episodes of PTLD. Collectively, these results show that the quantification of EBV DNA in EBV-related lymphoid malignancies is a valid tumor biomarker and a valuable adjunct in disease monitoring.
If EBV DNA reflected tumor load, it might reasonably be expected to be prognostically relevant. To evaluate this assumption, we used the median presentation EBV DNA of the whole group of EBV-related lymphomas as a cutoff. Despite the limitation that this series comprised a mixture of lymphomas with different histopathology and treatment, cases with diagnostic EBV DNA above the median presentation level could still be shown to have a significantly inferior OS. A similar trend was shown for DFS, although owing to the heterogeneous patient population, statistical significance was not reached.
Therefore, to analyze a more homogeneous population, we further studied our cohort of patients with NK cell lymphoma, who formed the largest subgroup in this series. NK cell lymphomas are highly aggressive malignancies with a peculiar geographic distribution. They are very uncommon tumors, occurring predominantly in Asian and Central American patients, and are exceptionally rare in Western countries.4 NK cell lymphomas are almost universally associated with EBV infection in the tumor cells. Owing to its rarity, prognostic markers for this tumor have not been well defined. Furthermore, NK cell lymphomas can be divided clinically into nasal, nonnasal, and disseminated subtypes,4 and it is undefined if each subtype may have different prognostic factors. We showed recently, for nasal NK cell lymphomas, that the IPI was the most significant factor in predicting CR rate and OS.26 In this study, with one of the largest series of patients with NK cell lymphomas reported to date, we showed that for all 3 clinical subtypes, the presentation EBV DNA was the most significant prognostic factor for disease-free survival (DFS). It was also one of the significant prognostic factors in univariate analysis of OS for all 3 subtypes. Furthermore, analysis of EBV DNA during treatment showed that a rise of EBV DNA that exceeded the presentation level, and failure to attain an undetectable EBV DNA, were also associated with an inferior OS. Biologically, the high-presentation EBV DNA might mean a high tumor load, the rise of EBV DNA a progression of disease, and the failure to attain undetectable EBV DNA an absence of a complete response. Therefore, EBV DNA provides an important means for prognostication of EBV-positive lymphomas at diagnosis and during treatment. This has important implications for the management of these patients. NK cell lymphomas have a poor treatment outcome, even with combined chemotherapy and radiotherapy.27-29 We have shown that the use of autologous HSCT might be beneficial to selected patients with NK cell lymphomas.30 Our observations suggest that EBV DNA may be used as a stratification factor at presentation and during treatment to identify bad-risk patients who may benefit from early high-dose chemotherapy and autologous HSCT. Accordingly, we are now conducting a prospective study to validate this proposition.
Quantification of EBV DNA by Q-PCR is highly sensitive and can be conveniently performed in the peripheral blood. A current problem of this assay is the use of various sources of EBV, including cell lines and plasmids, as standards for quantification,14-17 which might affect the assay in different ways. EBV-positive cell lines have been used as the starting standard, with the EBV copy number per cell enumerated either by electron microscopy16,17 or previous in situ hybridization.14 Others have used EBV DNA fragments in competitive PCR methods.15 Apart from affecting the actual numerical values of the assays, cellular or plasmid DNA might have different amplification efficiencies. Consequently, the reported EBV DNA, quantified with comparable technologies from patients with similar diseases, might appear to differ by several logs in different studies.14-17 Although this spurious difference because of the use of different standards hinders a direct comparison of the numerical results of these studies, the biologic phenomena that they showed were nevertheless consistent. Therefore, for this test to be comparable and more extensively validated, an agreed EBV standard is ultimately needed. Notwithstanding this limitation, EBV DNA is potentially useful in the diagnosis and follow-up of patients with EBV-related lymphomas, as well as in the assessment of the efficacies of chemotherapeutic regimens and monoclonal antibodies such as rituximab in the treatment for these tumors. It may be particularly helpful in lymphomas in which clonal markers evaluable by PCR are not available. These include NK cell lymphomas and Hodgkin lymphoma, whereby clonal rearrangement of the immunoglobulin and T-cell receptor genes could not be applied for detection of minimal residual disease. In NK cell lymphomas in which zonal necrosis is a prominent feature, the definition of neoplastic cells, particularly in re-biopsies after treatment, is often challenging even to the experienced histopathologist.31 In Hodgkin lymphoma, the early detection of relapse and the distinction between residual tumor or fibrotic lymph nodes is difficult.32 Preliminary data in a small number of patients with Hodgkin lymphomas have shown that quantification of EBV DNA might be useful in these situations.33,34 Finally, prospective studies are needed to validate the role of EBV DNA testing in the management algorithms of EBV-positive lymphomas, as has been defined for patients with PTLD.16,17
Prepublished online as Blood First Edition Paper, March 18, 2004; DOI 10.1182/blood-2003-12-4197.
Supported by the Kadoorie Charitable Foundation and Committee for Research and Conference (grant 10204377).
W.Y.A. and A.P. contributed equally to this work.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.