Abstract
Several lines of investigation suggest that granulocyte colony-stimulating factor (G-CSF) augments all-trans retinoic acid (ATRA)–induced neutrophil differentiation in acute promyelocytic leukemia (APL). We sought to characterize the relationship between G-CSF– and ATRA-mediated neutrophil differentiation. We established a G-CSF receptor-transduced promyelocytic cell line, EPRO-Gr, derived from the granulocyte-macrophage colony-stimulating factor (GM-CSF)–dependent EPRO cell line harboring a dominant-negative retinoic acid receptor alpha (RARα). In EPRO-Gr, neutrophil differentiation occurs either in GM-CSF upon addition of ATRA or upon induction with G-CSF alone. Transient transfection of EPRO-Gr cells with a RARE-containing reporter plasmid demonstrates increased activity in the presence of ATRA, but not G-CSF, while STAT3 phosphorylation occurs only in response to G-CSF. This suggests that ATRA-mediated differentiation of EPRO-Gr cells occurs via a RARE-dependent, STAT3-independent pathway, while G-CSF–mediated differentiation occurs via a RARE-independent, STAT3-dependent pathway. ATRA and G-CSF thus regulate differentiation by divergent pathways. We characterized these pathways in the APL cell line, NB4. ATRA induction of NB4 cells resulted in morphologic differentiation and up-regulation of C/EBPϵ and G-CSFR, but not in STAT3 phosphorylation. The addition of G-CSF with ATRA during NB4 induction resulted in STAT3 phosphorylation but did not enhance differentiation. These results may elucidate how G-CSF and ATRA affect the differentiation of primary and ATRA-resistant APL cells.
Introduction
Granulopoiesis involves the coordinated action of cytokines and transcription factors (reviewed in Friedman1 and Lotem and Sachs2 ). Of the cytokines affecting neutrophil development, granulocyte colony-stimulating factor (G-CSF) plays an essential role in proliferation, survival, and differentiation of granulocyte precursors in the marrow. G-CSF acts by binding to its receptor (G-CSFR), a member of the class I cytokine receptor superfamily expressed on myeloid progenitors (reviewed in Avalos3 ). Ligand binding to the G-CSFR causes phosphorylation of intracellular tyrosines and activation of signal transduction pathways, including Ras/Raf/MAPK, PI3-kinase, and JAK/STAT cascades (reviewed in Avalos3 ). Of these pathways, a direct differentiation signal appears to be mediated by activation of the signal transducer and activator of transcription 3 (STAT3) protein.4 Despite increased understanding of G-CSF signaling pathways, their function during G-CSF–induced differentiation remains unclear. Furthermore, mice lacking both G-CSF and G-CSFR can produce morphologically mature neutrophils, albeit at lower than normal levels,5,6 suggesting that neutrophil development can proceed via G-CSF–dependent and –independent pathways.
Retinoic acid also promotes the differentiation of normal and leukemic myeloid cells (reviewed in Kastner and Chan7 and Zelent et al8 ). A role for the vitamin A derivative all trans retinoic acid (ATRA) in neutrophil maturation is implied by studies of acute promyelocytic leukemia (APL). In all cases of APL, the retinoic acid receptor (RARα) gene is the target of chromosomal rearrangements. The resulting fusion proteins possess a variable N-terminus derived from the PML, PLZF, NuMA, nucleophosmin, or STAT5b genes (reviewed in Zelent et al8 ) and a constant C-terminus contributed by part of the RARα gene, including its DNA- and ligand-binding domains. This suggests that perturbation of normal RARα function is critical to the pathogenesis of APL.
The most common APL translocation, t(15;17), results in expression of a PML-RARα fusion product that is hypothesized to be a dominant-negative repressor of retinoic acid target genes, resulting in a block to terminal neutrophil differentiation. Superphysiologic levels of ATRA are thought to overcome the block to RA signaling in APL cells and restore expression of retinoic acid target genes (Zelent et al8 and references therein). An alternative hypothesis has suggested that differentiation is indirectly disrupted by the PML-RARα fusion protein by sequestration of myeloid transcriptional activators, such as C/EBPα.9
Several studies suggest that G-CSF–induced signaling might cooperate with ATRA-induced pathways to control myeloid differentiation. For example, in vitro studies have suggested that differentiation of APL cells by ATRA is enhanced by G-CSF.10-12 More recent evidence suggests that a combination of G-CSF and ATRA induces differentiation of APL cells unresponsive to ATRA alone.13,14 Finally, it has been reported that an APL patient expressing the ATRA-resistant translocation t(11;17) (PLZF-RARα) achieved complete remission upon the addition of G-CSF to the ATRA-induction regimen.15
It is likely that both G-CSF and RA stimulate common downstream transcription factors regulating neutrophil development. Two transcription factors belonging to the CCAAT/enhancer-binding protein (C/EBP) family, C/EBPα and C/EBPϵ, are key regulators of myeloid differentiation. C/EBPα knockout mice reveal impaired myelopoiesis with a differentiation arrest at an early myeloid progenitor stage.16 G-CSF receptor expression is impaired in these mice, and ectopic G-CSF receptor expression can rescue the differentiation of C/EBPα–/– progenitors. C/EBPϵ–/– mice also display abnormal myelopoiesis, producing functionally and morphologically abnormal granulocytes.17 C/EBPϵ has been shown to be directly induced by retinoic acid at the mRNA level.18 We and others have demonstrated recently that C/EBPϵ overexpression in murine 32Dcl3 cells bypasses the requirement for G-CSF in the induction of terminal neutrophil differentiation.19,20 These observations suggest that myelopoiesis proceeds via the sequential activation/expression of C/EBPα, C/EBPϵ, and G-CSFR.
In this study, we used 2 different models to further characterize ATRA- and G-CSF–mediated differentiation of promyelocytes. First, we transduced the GM-CSF–dependent EPRO cell line, which is blocked at the promyelocyte stage due to expression of a dominant-negative retinoic acid receptor alpha gene (RARα403),21 with a retrovirus that provides constitutive overexpression of the murine G-CSFR gene. The resulting EPRO-Gr cells were then shown to undergo terminal neutrophil differentiation in response to either ATRA (≥ 1 μM) or G-CSF. We then evaluated ATRA and G-CSF–induced responses in the APL cell line, NB4.22 We demonstrate that G-CSF but not ATRA-mediated signaling results in phosphorylation of STAT3, whereas ATRA but not G-CSF activates a reporter gene driven by retinoic acid receptor elements (RAREs). Additionally, ATRA induction of NB4 cells leads to indirect up-regulation of G-CSFR gene expression but not to STAT3 phosphorylation. Induction of ATRA-primed NB4 cells with G-CSF, however, did lead to STAT3 phosphorylation. These results suggest that up-regulation of G-CSFR by ATRA might mediate the synergistic effects of G-CSF on ATRA treated primary APL cells. However, G-CSF induction appears to have little effect on the maturation of ATRA-primed NB4 cells.
Materials and methods
Cell lines
EML/EPRO cells. EML cells were provided by Dr Schickwann Tsai21 (Mt Sinai School of Medicine, New York, NY). EPRO (EML derived Promyelocytes) cells were generated from EML cells by treatment with interleukin-3 (IL-3) (5% WeHi conditioned media) and ATRA (10 μM; Sigma, St Louis, MO).21 EPRO cells were maintained in Iscove modified Dulbecco medium (IMDM), 20% horse serum, recombinant GM-CSF (10 ng/mL), and penicillin/streptomycin as described by Tsai et al,21 or in minimal serum conditions in AIM V (Gemini Bio-Products, Woodland, CA), 1%-5% fetal calf serum, recombinant murine GM-CSF at 10 ng/mL(Amgen, Thousand Oaks, CA), and penicillin/streptomycin.
NB4 cells. NB4 cells were maintained as previously described.22
Generation of murine wild-type G-CSFR expressing EPRO cells
A vector containing the full-length murine G-CSF receptor cDNA (mWT G-CSFR), pEMCV.SRα-mGCSFR, was provided by Dr Dan Link, Washington University, St Louis, MO. The cDNA was excised by digestion with XhoI, and cloned into the retroviral vector pBABE-Puro23 at a SalI site. BOSC cells were transiently transfected with the pBABE-Puro-mWT G-CSFR plasmid or vector alone using LipofectAMINE (Gibco BRL, Grand Island, NY). After overnight incubation at 37°C in 5% CO2, transfected BOSC cells were overlaid with EML cells in standard growth media at a density of 0.5 × 106 cells/mL. Following 24 hours coculture, EML cells were harvested, expanded in growth media, and selected in 1.0 μg/mL puromycin (Sigma). Transduced EML cells were differentiated to the EPRO stage as previously described.21
In vitro proliferation and differentiation
Proliferating EPRO-Gr, NB4, and APL cells were harvested and washed twice with phosphate-buffered saline (PBS) and resuspended at 2-5 × 105 cells/mL in either standard serum or minimal serum media in the presence of G-CSF (100 ng/mL; Amgen) and/or ATRA (10 μM for EPRO-Gr and APL cells, 5 μM for NB4 cells; Sigma). Cell density was maintained less than or equal to 1.0 × 106 cells/mL by dilution with fresh medium. When dilution was not required, partial medium exchange was performed every 2 days. Phenotypic maturation was determined by Wright-Giemsa staining of cytospin smears.
Northern analysis
RNA was extracted during induction of EPRO-Gr, NB4, and APL cells using the TRIzol reagent (Life Technologies, Rockville, MD) according to manufacturer's specification. Total RNA (10 μg) was analyzed by Northern blot analysis and probed with designated 32P-dCTP–labeled cDNA probes as described previously.19,24 Hybridizations were performed either overnight at 42°C in the presence of 50% formamide or for 2 hours in ExpressHyb (Sigma). Hybridization filters were washed with 2 × SSC (300 mM NaCl and 60 mM sodium citrate)/0.1% sodium dodecyl sulfate (SDS) twice at room temperature for 10 minutes, followed by 2 washes in 0.1 × SSC (15 mM NaCl and 3 mM sodium citrate)/0.1% SDS at 55°C to 60°C for 15 minutes. Blots were autoradiographed at –70°C with an intensifying screen.
Luciferase reporter analysis
A plasmid (pBStkRARE-luc) bearing 3 retinoic acid responsive elements (RAREs) cloned upstream of the luciferase reporter gene was provided by Dr Ron Evans (Salk Institute, La Jolla, CA). Transient transfection was carried out in EPRO-Gr cells as follows. Approximately 1 × 107 cells were pelleted at 1000 × g, washed twice with 1 × PBS, and resuspended in 180 μL HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffered saline in 0.4 mm electroporation cuvettes (BioRad, Hercules, CA). Added were 20 μg pBStkRARE-luc plasmid and 2 μg pCMVβ-gal (Clontech, Palo Alto, CA), a control plasmid used to monitor transfection efficiency. The DNA cell samples were electroporated using a Biorad Gene Pulser (BioRad) at 300 V with 500 μF capacitance. Transiently transfected cells were incubated at 37°C, 5% CO2 for 16-20 hours in induction media as described above. Luciferase activity was determined using a kit from Promega Biotech (Madison, WI), per manufacturer's instructions. Luciferase expression levels were normalized to β-galactosidase expression and reported as fold increase in luciferase activity over pBStkRARE-luc as previously described.25
Western blot analysis
Uninduced and induced EPRO-Gr and NB4 cells were washed once with 1 × PBS and cell pellets lysed in 2 × gel sample buffer (125 mM Tris [tris(hydroxymethyl)aminomethane], pH 6.8; 4% SDS; 10% glycerol; 2 mg/mL bromophenol blue; 57 mM β-mercaptoethanol). Total protein lysates and molecular weight markers (Rainbow; Amersham Pharmacia Biotech, Piscataway, NJ) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using either 7% (BioRad) or 4% to 12% (Invitrogen, Carlsbad, CA) precast gels. Gels were transferred onto polyvinylidenefluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, MA) using a tank transfer system (BioRad) in transfer buffer (25 mM Tris, 192 mM glycine, 20% MeOH, 0.1% SDS). EPRO-Gr blots were probed with polyclonal anti–murine G-CSFR (M-20; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:800 dilution in blocking buffer (TBS-Tween: 20 mM Tris, pH 7.5; 150 mM NaCl; 0.1% Tween 20; 5% nonfat dry milk). NB4 cells were probed with anti–human G-CSFR antibody (BD Biosciences, San Diego, CA) at a dilution of 1:500 in blocking buffer. Antibodies to specific STAT proteins were diluted in blocking buffer at appropriate concentrations: rabbit polyclonal IgG anti-STAT3: 1:1000 (Cell Signaling Technology, Beverly, MA), mouse monoclonal anti-STAT1: 1:2500 (S21120, Transduction Laboratories, Lexington, KY), and mouse monoclonal anti-STAT5: 1:250 (610191, Transduction Laboratories).
STAT activation was analyzed using rabbit polyclonal IgG anti–phospho-STAT3 (Tyr 705), anti–phospho-STAT5 (Tyr 694), and anti–phospho-STAT1 (Tyr 701) antibodies (Cell Signaling Technology), each diluted 1:1000 in blocking buffer. All blots were exposed to primary antibodies overnight at 4°C and washed with TBS-Tween-20. Blots were probed with horseradish peroxidase (HRP)–linked anti–rabbit IgG antibodies (Cell Signaling Technology) and HRP-linked anti–mouse IgG antibodies (Amersham Pharmacia Biotech), diluted 1:2000 in blocking buffer for 1 hour at room temperature. Chemiluminescent detection was performed according to manufacturer's recommendations (Phototope-HRP Western Blot Detection System; Cell Signaling Technology).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed using the protocol for the acetyl-histone H4 ChIP Assay Kit (Upstate Biotechnology, Lake Placid, NY) as previously described.26 Polymerase chain reaction (PCR) was performed using 1 μL ChIP DNA using oligos flanking the C/EBP binding site within the G-CSFR promoter27 as follows: G-CSFR forward primer (FP): 5′-TCACCAGCTTCCCTCACAGG-3′; G-CSFR reverse primer (RP): 5′-GCTGCAGTCCAGCTTCTCTC-3′. PCR products were subcloned into the pCRII vector (Invitrogen) and sequenced by standard dideoxy sequencing technology to confirm their identity.
G-CSFR surface expression
Surface G-CSFR expression in EPRO-Gr and NB4 cells was determined by a modification of the technique described for GM-CSFR.28 Briefly, 10 μgof human G-CSF (Amgen) in 0.1 M sodium bicarbonate buffer pH 8.4, containing 0.02% (wt/vol) Tween-20, was incubated with a 300-fold molar excess of NHS-LC-Biotin (Pierce, Rockford, IL) for 2 hours at 20°C. Unincorporated biotin was removed by size exclusion filtration using Microcon 30 spin columns (Amicon, Beverly, MA). Filter columns were equilibrated with 5% Triton X-100, and the biotin–G-CSF reaction was added and spun at 1000 × g for 15 minutes. Activity of the biotin-labeled G-CSF was verified by the ability to support survival of EPRO-Gr cells. Preparation of cells was essentially as described,28 followed by incubation with 100 ng streptavidin-phycoerythrin (PE) (cat. no. 554061; PharMingen, San Diego, CA) for 30 minutes on ice. Cells were washed twice with HFN (Hanks balanced salt solution [Gibco] with 2% [vol/vol] fetal bovine serum [Gibco] and 0.05% sodium azide), resuspended in 200 μL HFN, and analyzed by flow cytometry using a FACS Vantage flow cytometer (Becton Dickinson, San Jose, CA) with CellQuest software. As a control for EPRO-Gr, non–streptavidin–PE-labeled cells were analyzed in tandem. For NB4, non–streptavidin–PE-labeled cells and cells labeled with streptavidin-PE in the absence of biotin–G-CSF were used as controls.
Results
EPRO cells lack expression of the G-CSF receptor and fail to respond to G-CSF–induced survival and differentiation
EML is a stem cell factor (SCF)–dependent line that is blocked in myeloid differentiation by a dominant-negative retinoic acid receptor alpha gene (RARα403). A GM-CSF–dependent promyelocytic cell line EPRO (EML derived Promyelocytes) can be generated from EML cells upon exposure to IL-3 and ATRA.21 EPRO cells in turn undergo terminal neutrophil differentiation upon addition of pharmacologic doses of ATRA. We initially evaluated the G-CSF responsiveness of the EPRO cell line. We transferred proliferating EPRO cells maintained in GM-CSF to G-CSF–containing medium lacking GM-CSF. Following 12 hours of incubation, fewer than 5% of the EPRO cells were viable by trypan-blue exclusion (data not shown), suggesting that EPRO cells depend on GM-CSF for survival and do not respond to G-CSF. We then performed Northern analysis to determine whether the G-CSF receptor is expressed in EPRO cells. Uninduced EPRO cells express very low levels of G-CSFR mRNA; expression is up-regulated after 24 hours of ATRA induction, which coincides with up-regulated expression of lactoferrin.29 Although ATRA-primed EPRO cells showed some G-CSF responsiveness (data not shown), the need to treat with ATRA made it difficult to separate retinoid signaling and G-CSF signaling; therefore, we proceeded to make a transduced G-CSF–expressing cell line.
Generation of EPRO cells overexpressing murine WT G-CSFR (EPRO-Gr)
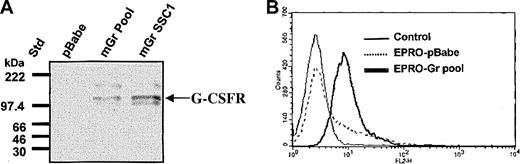
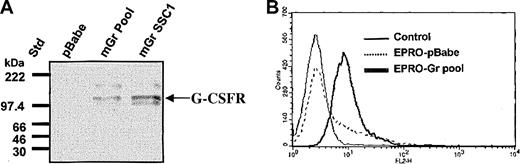
The murine G-CSFR gene was retrovirally transduced into EML cells and the GCSFR+ cells were then used to derive GM-CSF–dependent EPRO cells (EPRO-Gr) ectopically expressing the G-CSFR (“Materials and methods”). EPRO-Gr single cell clones were derived by limiting dilution of puromycin-selected cells. Control cells expressing the empty pBabe-Puro vector (EPRO-pBABE) were generated in parallel. Overexpression of the murine G-CSFR cDNA by EPRO-Gr was confirmed by Western blot analysis (Figure 1A). G-CSFR protein is detectable in lysates derived from both the pooled EPRO-Gr cells (mGr Pool, lane 2) and a representative single cell clone of EPRO-Gr cells (mGr SSC1, lane 3), but not in EPRO cells expressing the vector alone (pBabe, lane 1). Eight single cell clones analyzed for G-CSFR overexpression were positive by Western blot analysis, with little variation in expression between clones (data not shown).
Expression of murine G-CSFR in parental EPRO cells and EPRO-Gr cells. Wild-type murine G-CSFR cDNA was cloned into the retroviral vector pBabe-Puro, and the construct was transduced into EML cells. EML-Gr cells then were induced to generate EPRO cells overexpressing the G-CSFR (EPRO-Gr), which were then analyzed for total G-CSFR protein expression (A) and cell surface G-CSFR expression (B). (A) Cell lysates (5 × 103 cells/μL2 × GSB) were harvested from EPRO-Gr pools, EPRO-Gr single-cell clones, and EPRO-pBabe (vector alone). Lysates were subjected to 7% SDS-PAGE and Western analysis probing with rabbit polyclonal anti–murine G-CSF receptor. (B) Recombinant human G-CSF was biotinylated at a molar ratio of 300:1 (biotin/G-CSF) and incubated with EPRO-Gr cells at a final G-CSF concentration of 26 nM. Surface expression of G-CSFR was then determined by flow cytometry using phycoerythrin-labeled streptavidin. EPRO-pBabe cells were analyzed in tandem, and nonstreptavidin–PE-labeled cells were used as control.
Expression of murine G-CSFR in parental EPRO cells and EPRO-Gr cells. Wild-type murine G-CSFR cDNA was cloned into the retroviral vector pBabe-Puro, and the construct was transduced into EML cells. EML-Gr cells then were induced to generate EPRO cells overexpressing the G-CSFR (EPRO-Gr), which were then analyzed for total G-CSFR protein expression (A) and cell surface G-CSFR expression (B). (A) Cell lysates (5 × 103 cells/μL2 × GSB) were harvested from EPRO-Gr pools, EPRO-Gr single-cell clones, and EPRO-pBabe (vector alone). Lysates were subjected to 7% SDS-PAGE and Western analysis probing with rabbit polyclonal anti–murine G-CSF receptor. (B) Recombinant human G-CSF was biotinylated at a molar ratio of 300:1 (biotin/G-CSF) and incubated with EPRO-Gr cells at a final G-CSF concentration of 26 nM. Surface expression of G-CSFR was then determined by flow cytometry using phycoerythrin-labeled streptavidin. EPRO-pBabe cells were analyzed in tandem, and nonstreptavidin–PE-labeled cells were used as control.
Cell surface expression of G-CSFR in EPRO-Gr cells was confirmed by flow cytometry. Detection of surface binding of biotinylated G-CSF was assessed using phycoerythrin-labeled streptavidin (Figure 1B). EPRO-pBABE cells displayed low level endogenous G-CSFR expression (< 20%) as predicted by the minimal G-CSFR mRNA observed by northern analysis (data not shown). In contrast, flow cytometric analysis of EPRO-Gr cells revealed uniformly high fluorescence, suggesting high levels of surface G-CSFR expression.
Neutrophil differentiation of EPRO-Gr cells
EPRO-Gr and EPRO-pBabe cells maintained in GM-CSF under standard growth conditions21 demonstrate a low level (0%-10%) of spontaneous morphologic neutrophil differentiation, a level comparable to that seen in untransduced EPRO cells.29 We have previously shown that spontaneous neutrophil differentiation is minimized when EPRO cells are maintained in minimal serum (1.0%-5.0% fetal calf serum).29
We verified the ability of EPRO-Gr cells to undergo neutrophil differentiation. As expected, induction of uninduced EPRO-Gr cells (Figure 2A, u) with ATRA (10 μM) in the presence of GM-CSF resulted in decreased proliferation (data not shown), followed by appearance of morphologically mature neutrophils (Figure 2A, a). These changes were identical to those observed in EPRO cells derived from the parent EML cell line.29
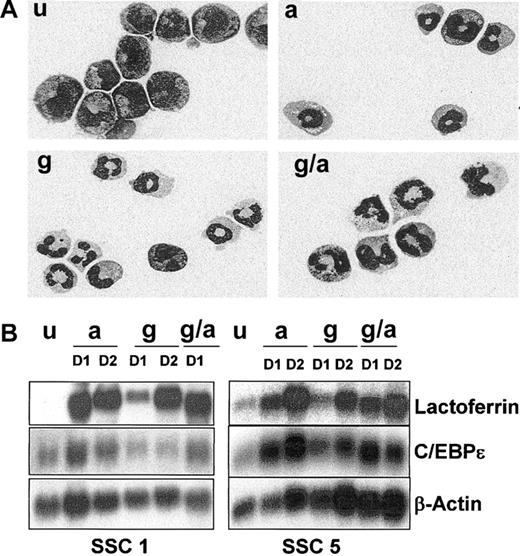
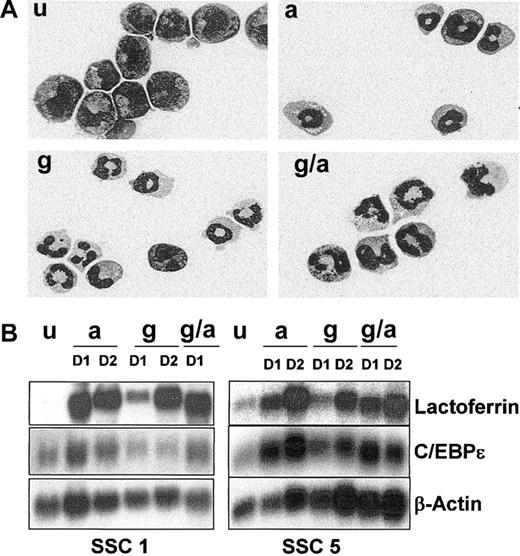
Neutrophil differentiation of EPRO-Gr cells. The effects of G-CSF (100 ng/mL) and/or ATRA (10 μM) on differentiation of EPRO-Gr was monitored by Wright-Giemsa stained cytospins and northern analysis. EPRO-Gr cells (u) were induced by the addition of ATRA (a), G-CSF-alone (g), or the combination of G-CSF plus ATRA (g/a). Cells were harvested daily. (A) Wright-Giemsa–stained cytospins were subjected to morphologic assessment by light microscopy. Shown are representative cells induced for 3 days as described above (original magnification, × 100). (B) Total RNA was isolated from EPRO-Gr cells induced as described above and subjected to Northern analysis. RNA was isolated from uninduced EPRO-Gr and cells induced for 24 hours (D1) and 48 hours (D2). Ten micrograms total RNA from each sample was analyzed by Northern blotting. Blots were sequentially probed with 32P-labeled cDNA probes for mouse lactoferrin and human C/EBPϵ. To monitor loading of RNA, the blot was probed with mouse β-actin.
Neutrophil differentiation of EPRO-Gr cells. The effects of G-CSF (100 ng/mL) and/or ATRA (10 μM) on differentiation of EPRO-Gr was monitored by Wright-Giemsa stained cytospins and northern analysis. EPRO-Gr cells (u) were induced by the addition of ATRA (a), G-CSF-alone (g), or the combination of G-CSF plus ATRA (g/a). Cells were harvested daily. (A) Wright-Giemsa–stained cytospins were subjected to morphologic assessment by light microscopy. Shown are representative cells induced for 3 days as described above (original magnification, × 100). (B) Total RNA was isolated from EPRO-Gr cells induced as described above and subjected to Northern analysis. RNA was isolated from uninduced EPRO-Gr and cells induced for 24 hours (D1) and 48 hours (D2). Ten micrograms total RNA from each sample was analyzed by Northern blotting. Blots were sequentially probed with 32P-labeled cDNA probes for mouse lactoferrin and human C/EBPϵ. To monitor loading of RNA, the blot was probed with mouse β-actin.
We next examined the effect of G-CSF on EPRO-Gr cells. EPRO-Gr cells were removed from GM-CSF containing growth medium, washed, and transferred to G-CSF containing medium. G-CSF induction resulted in decreased proliferation similar to that observed with ATRA induction, and Wright-Giemsa staining revealed more than 95% of G-CSF–induced cells exhibited mature neutrophil morphology (Figure 2A, g). The addition of ATRA to G-CSF did not alter the kinetics of decreased proliferation (data not shown) nor did it affect the extent of neutrophil maturation when compared with G-CSF alone (Figure 2A, g/a).
We have previously demonstrated that ATRA-induced differentiation of factor-dependent cell lines such as EML, MPRO, and EPRO results in late neutrophil-specific gene expression, including the expression of secondary granule protein (SGP) genes.29 To determine whether G-CSF induction also resulted in a normal pattern of late gene expression, lactoferrin (LF) and C/EBPϵ gene expression was analyzed by Northern blot analysis of 2 EPRO-Gr single cell clones induced with ATRA, G-CSF, and ATRA plus G-CSF. By day 1 (D1) of induction, LF mRNA levels were up-regulated in the presence of either ATRA, G-CSF, or ATRA plus G-CSF (Figure 2B, levels range from 4- to 10-fold above levels in uninduced cells as determined by image densitometry [Image-QuaNT software]). Levels of expression were comparable by day 2 (D2) in all induction regimens employed (A, G and G/A). C/EBPϵ mRNA was detectable in uninduced EPRO-Gr SSC cells and was up-regulated in the presence of ATRA or ATRA plus G-CSF by day 1 (Figure 2B, levels range from 2- to 3-fold above levels in uninduced cells). However, while C/EBPϵ is expressed in both clones in the presence of G-CSF, its expression does not appear to increase by 2 days of induction, and levels do not increase upon further incubation in G-CSF (data not shown, relative intensities up to 3 days of induction are similar to that expressed in uninduced cells as determined by densitometry analysis).
Retinoic acid response element (RARE)–luciferase reporter assay of EPRO-Gr
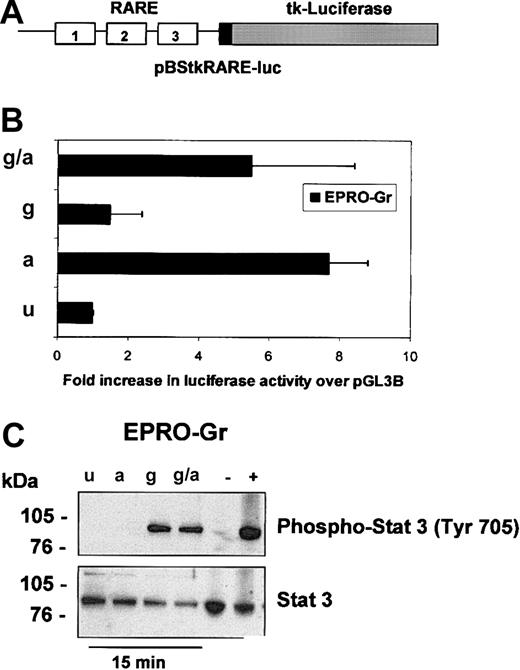
The biologic effects of retinoic acid (RA) are mediated by binding to RA receptors (RARs). The ligand-receptor complex regulates RA target genes by interaction with retinoic acid responsive elements (RAREs) present in their promoters (reviewed in Melnick and Licht30 ). Overexpression of dominant-negative forms of RARα in myeloid cells inhibits normal granulopoiesis,21,31 an effect reversible by pharmacological doses of ATRA. This ATRA-induced reversal of the block to differentiation in these cells is attributed to the transactivation of RA-responsive genes.32 We therefore examined whether neutrophil differentiation of EPRO-Gr is associated with RARE transactivation. EPRO-Gr cells were transiently transfected with a RARE-containing reporter gene plasmid, pBStkRARE-luc (Figure 3A). Induction of transfected cells with ATRA resulted in a 7.7-fold increase in luciferase activity (Figure 3B, a) over uninduced cells (Figure 3B, u). G-CSF induction of transfected EPRO-Gr cells (Figure 3B, g) did not significantly (1.5-fold) alter luciferase activity over control levels (Figure 3B, u). Furthermore, the addition of G-CSF to ATRA did not increase transactivation of pBStkRARE-luc over that observed with ATRA alone (5.5-fold, Figure 3B, g/a). These data suggest that activation of a RARE is dependent only on ATRA and is not affected by activation of the G-CSF receptor.
Analysis of retinoic acid response element (RARE)–luciferase reporter gene activation and STAT3 phosphorylation in EPRO-Gr inductions. Induced EPRO-Gr cells were analyzed for activation of RAREs (A,B) by transient transfections with pBStkRARE-luc and treatment of cells with various induction media as outlined in Figure 2 (u-uninduced; a-ATRA; g-GCSF; g/a-GCSF plus ATRA) and for activation of STAT3 (C) under the same conditions. (A) Shown is a diagram of the pBStkRARE luciferase reporter construct containing 3 RAREs placed in series upstream of the 158-bp tk promoter fragment and luciferase (“Materials and methods”). (B) Cell lysates were obtained 24 hours after transfection, and luciferase activity was measured. A beta-galactosidase expression vector was cotransfected at a ratio of 20:1 to normalize for transfection efficiency. Luciferase activity in EPRO-Gr cells is reported as fold increase in relative light units (RLU) above pBStkRARE-luc in control medium (GM-SCF). The figure represents the mean ± SE value from 3 independent experiments, each performed in duplicate. (C) Following 60 minutes of serum starvation, total cell lysates (5 × 103 cells/μL2 × GSB) were harvested from uninduced (u) EPRO-Gr cells, and cells induced with ATRA (a), G-CSF (g), and G-CSF plus ATRA (g/a) for 15 minutes. Lysates were subjected to 4%-12% SDS-PAGE. Western analysis was subsequently performed and probed sequentially with rabbit-polyclonal IgG anti–phospho-STAT3 (Tyr 705) and rabbit-polyclonal IgG anti-STAT3 as primary antibodies. Binding of primary antibodies was detected with HRP-linked anti–rabbit IgG antibodies and chemiluminescent detection. As controls, equivalent amounts of lysates from HeLa cells treated with INF-α (positive control; 100 ng/mL) and untreated HeLa cells (negative control) were run in parallel.
Analysis of retinoic acid response element (RARE)–luciferase reporter gene activation and STAT3 phosphorylation in EPRO-Gr inductions. Induced EPRO-Gr cells were analyzed for activation of RAREs (A,B) by transient transfections with pBStkRARE-luc and treatment of cells with various induction media as outlined in Figure 2 (u-uninduced; a-ATRA; g-GCSF; g/a-GCSF plus ATRA) and for activation of STAT3 (C) under the same conditions. (A) Shown is a diagram of the pBStkRARE luciferase reporter construct containing 3 RAREs placed in series upstream of the 158-bp tk promoter fragment and luciferase (“Materials and methods”). (B) Cell lysates were obtained 24 hours after transfection, and luciferase activity was measured. A beta-galactosidase expression vector was cotransfected at a ratio of 20:1 to normalize for transfection efficiency. Luciferase activity in EPRO-Gr cells is reported as fold increase in relative light units (RLU) above pBStkRARE-luc in control medium (GM-SCF). The figure represents the mean ± SE value from 3 independent experiments, each performed in duplicate. (C) Following 60 minutes of serum starvation, total cell lysates (5 × 103 cells/μL2 × GSB) were harvested from uninduced (u) EPRO-Gr cells, and cells induced with ATRA (a), G-CSF (g), and G-CSF plus ATRA (g/a) for 15 minutes. Lysates were subjected to 4%-12% SDS-PAGE. Western analysis was subsequently performed and probed sequentially with rabbit-polyclonal IgG anti–phospho-STAT3 (Tyr 705) and rabbit-polyclonal IgG anti-STAT3 as primary antibodies. Binding of primary antibodies was detected with HRP-linked anti–rabbit IgG antibodies and chemiluminescent detection. As controls, equivalent amounts of lysates from HeLa cells treated with INF-α (positive control; 100 ng/mL) and untreated HeLa cells (negative control) were run in parallel.
STAT3 phosphorylation in EPRO-Gr cells
Current evidence suggests an important role for transcriptional activation by phosphorylated signal transduction and activator of transcription 3 (STAT3) in G-CSF–induced neutrophil differentiation (reviewed in Coffer et al33 ). We therefore examined whether induction of EPRO-Gr cells is associated with STAT3 phosphorylation. Western blot analyses of EPRO-Gr cells were performed using antibodies specific for STAT3 and phosphorylated-STAT3 (Tyr 705). EPRO-Gr cells were induced with ATRA, G-CSF, and G-CSF plus ATRA and cell lysates recovered 15 minutes after induction. Uninduced EPRO-Gr cells expressed only unphosphorylated STAT3 (Figure 3C, u, lower panel). G-CSF induction of EPRO-Gr cells resulted in rapid STAT3 phosphorylation (Figure 3C, g, upper panel). In contrast, cells induced with ATRA demonstrated no STAT3 phosphorylation (Figure 3C, a, top panel). Furthermore, the addition of ATRA to G-CSF did not augment STAT3 phosphorylation over G-CSF alone (Figure 3C, g/a, top panel). Total STAT3 protein levels remained unchanged during the 15-minute induction of the EPRO-Gr cells (Figure 3C, lower panel). Identical results were obtained for EPRO-Gr cells induced for 90 minutes (data not shown).
The levels of STAT1 and STAT5, both known to play a role in myeloid differentiation,33 also were examined. While both proteins were present in these cells, STAT1 was not phosphorylated in uninduced or EPRO-Gr cells induced with G-CSF or ATRA (data not shown). STAT5 phosphorylation, however, was detectable in uninduced EPRO-Gr cells and was not modulated by any induction regimen (data not shown).
ATRA-induced gene expression in the acute promyelocytic cell line, NB4
The ability of G-CSF to induce maturation despite expression of a dominant negative RARα receptor in our EPRO-Gr cell model raised the question as to why G-CSF signaling cannot bypass the maturation block in APL cells. We therefore studied the APL cell line NB4, which harbors the t(15;17) translocation and undergoes morphologic maturation in response to ATRA.22 Previous studies have shown that addition of G-CSF to ATRA enhances the maturation of NB4 cells when compared to ATRA alone.10-12 However, we have previously shown that treatment of NB4 cells with G-CSF alone does not lead to neutrophil differentiation.34 This may reflect the low level of G-CSFR expression in uninduced NB4 cells. Importantly, expression of the G-CSF receptor is greatly increased in NB4 cells upon induction with ATRA.11,35 These data suggest APL cells may exhibit a certain level of responsiveness to G-CSF, but only after they have undergone priming with ATRA to up-regulate G-CSFR expression. We therefore determined whether ATRA-induced up-regulation of G-CSFR expression in NB4 cells leads to G-CSF responsiveness.
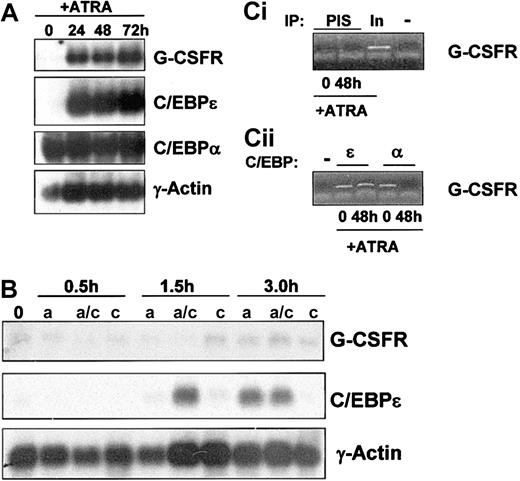
As previously reported, ATRA induction of NB4 cells resulted in up-regulation of the G-CSFR transcript after 24 hours (Figure 4A, top panel). This increased expression required more than 3 hours of induction (Figure 4B, top panel) and did not occur in the presence of the protein synthesis inhibitor cycloheximide, suggesting that G-CSFR is likely an indirect target of ATRA. The transcriptional activator C/EBPα has been reported to regulate G-CSFR expression during myeloid development.27 We found that C/EBPα is expressed in uninduced NB4 cells and does not appear to be modulated during ATRA induction (Figure 4A, C/EBPα panel). mRNA levels of C/EBPϵ were minimally detectable in uninduced NB4 cells and were rapidly up-regulated during ATRA induction (Figure 4A, C/EBPϵ panel). Up-regulation of C/EBPϵ mRNA by short-term ATRA exposure (1.5 hours and 3 hours) occurs in the presence of cycloheximide (Figure 4B), confirming, as previously reported,18 that C/EBPϵ is a direct transcriptional target of ATRA. Of note, although expression of C/EBPϵ mRNA was not observed in cells induced with ATRA alone for 1.5 hours, expression was observed by 1.5 hours in cells treated with ATRA plus cycloheximide. The mechanism for this up-regulation remains to be established.
G-CSFR gene expression in NB4 cells treated with ATRA. (A) Total RNA (10 μg/lane) was isolated from NB4 cells induced to differentiate with ATRA (5 μM) for 0, 24, 48, and 72 hours and subjected to Northern analysis. Blots were sequentially probed with 32P-labeled cDNA probes for human G-CSFR, human C/EBPϵ, and rat C/EBPα. To monitor equal loading of RNA in each lane, the blot was probed with human γ-actin. (B) Northern analysis was performed on NB4 cells after short term (0.5, 1.5, and 3.0 hours) exposure to ATRA (5 μM), in the presence (a/c) and absence (a) of cycloheximide (10 μg/mL). As a control, NB4 cells were exposed to cycloheximide alone (c) in parallel. Total RNA (10 μg/lane) was isolated and subjected to Northern analysis as in 4A. (C) Chromatin immunoprecipitation (ChIP) analysis of C/EBPϵ and C/EBPα binding to the G-CSFR promoter. ChIP was performed from uninduced (0) and ATRA-induced (48 hours) NB4 cells using antibodies specific for C/EBPϵ (ϵ) and C/EBPα (α; ii) or preimmune serum (PIS) and a no-antibody control (–; i). The precipitated chromatin was analyzed using primers specific for the G-CSFR gene C/EBP site. Total input DNA (In; 1:10 dilution) was used as a positive control (i).
G-CSFR gene expression in NB4 cells treated with ATRA. (A) Total RNA (10 μg/lane) was isolated from NB4 cells induced to differentiate with ATRA (5 μM) for 0, 24, 48, and 72 hours and subjected to Northern analysis. Blots were sequentially probed with 32P-labeled cDNA probes for human G-CSFR, human C/EBPϵ, and rat C/EBPα. To monitor equal loading of RNA in each lane, the blot was probed with human γ-actin. (B) Northern analysis was performed on NB4 cells after short term (0.5, 1.5, and 3.0 hours) exposure to ATRA (5 μM), in the presence (a/c) and absence (a) of cycloheximide (10 μg/mL). As a control, NB4 cells were exposed to cycloheximide alone (c) in parallel. Total RNA (10 μg/lane) was isolated and subjected to Northern analysis as in 4A. (C) Chromatin immunoprecipitation (ChIP) analysis of C/EBPϵ and C/EBPα binding to the G-CSFR promoter. ChIP was performed from uninduced (0) and ATRA-induced (48 hours) NB4 cells using antibodies specific for C/EBPϵ (ϵ) and C/EBPα (α; ii) or preimmune serum (PIS) and a no-antibody control (–; i). The precipitated chromatin was analyzed using primers specific for the G-CSFR gene C/EBP site. Total input DNA (In; 1:10 dilution) was used as a positive control (i).
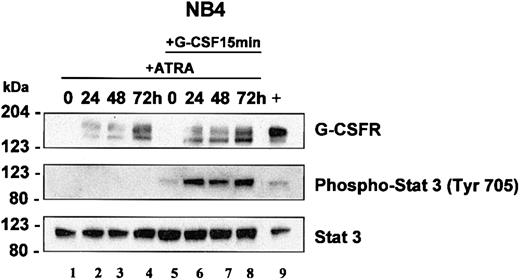
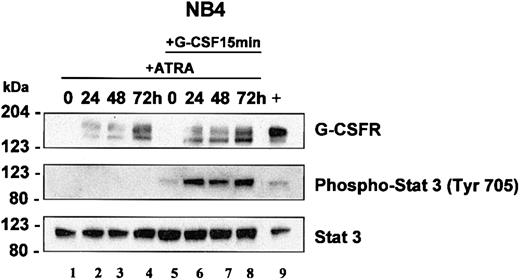
Western blot analysis of NB4 whole cell lysates showed similar results to our northern analysis. G-CSFR protein was undetectable in uninduced NB4 cells, but increased upon ATRA induction (Figure 5A, top panel, lanes 1-4). G-CSFR protein was readily detectable by 24 hours and remained elevated with continued exposure to ATRA up to 72 hours. The presence of 2 G-CSFR bands likely represents posttranslational modification of the protein.
STAT3 analysis in NB4 cells. NB4 cells were induced with ATRA (5 μM) for 0, 24, 48, and 72 hours. At each time interval, a representative sample was treated with G-CSF (100 ng/mL) for 15 minutes. Total cell lysates (5 × 103 cells/μL2 × GSB) from cells induced with ATRA alone (lanes 1-4) or treated with ATRA with subsequent treatment with G-CSF (lanes 5-8) were harvested and subjected to Western blot analysis. The blot was sequentially probed for STAT3, phospho-STAT3 (Tyr 705), and human G-CSFR as outlined in “Materials and methods.” As a positive control, lysates from EML cells overexpressing human G-CSFR were run in parallel (lane 9).
STAT3 analysis in NB4 cells. NB4 cells were induced with ATRA (5 μM) for 0, 24, 48, and 72 hours. At each time interval, a representative sample was treated with G-CSF (100 ng/mL) for 15 minutes. Total cell lysates (5 × 103 cells/μL2 × GSB) from cells induced with ATRA alone (lanes 1-4) or treated with ATRA with subsequent treatment with G-CSF (lanes 5-8) were harvested and subjected to Western blot analysis. The blot was sequentially probed for STAT3, phospho-STAT3 (Tyr 705), and human G-CSFR as outlined in “Materials and methods.” As a positive control, lysates from EML cells overexpressing human G-CSFR were run in parallel (lane 9).
Binding of C/EBP proteins to the G-CSFR promoter in NB4 cells
Abnormalities in C/EBPα protein levels36 or activity37 have been described in acute myeloid leukemia (AML). Although these abnormalities have not been observed in APL, it has been suggested that the PML/RARα gene product interacts with C/EBPα and inhibits its activity.9 Northern blot analysis indicates that uninduced and induced NB4 cells express abundant levels of the C/EBPα transcript (Figure 4A). We used chromatin immunoprecipitation (ChIP) to examine whether C/EBPα binds the G-CSFR promoter in NB4. Using oligonucleotides spanning the C/EBP binding site within the G-CSFR promoter, we performed ChIP analysis of uninduced and ATRA-induced NB4 cells and observed that C/EBPα binds the C/EBP site within the G-CSF promoter in uninduced cells (Figure 4Cii). Surprisingly, after ATRA induction, C/EBPα no longer binds the G-CSFR promoter. We also examined the ability of C/EBPϵ to bind to the G-CSFR promoter. Although C/EBPϵ mRNA is expressed at low levels, C/EBPϵ binds the G-CSFR promoter in uninduced NB4 cells, but unlike C/EBPα, binding persists in ATRA-induced cells (Figure 4Cii). No PCR product was obtained in ChIP analysis using preimmune serum (PIS) in uninduced or induced NB4 cells (Figure 4Ci), demonstrating the specificity of the C/EBPα and C/EBPϵ antibodies. Our findings indicate that both C/EBPα and C/EBPϵ are capable of binding the G-CSFR promoter in NB4 cells. However, how this binding may contribute to the expression of the G-CSFR gene remains to be further elucidated.
STAT3 phosphorylation in NB4 cells
In NB4 cells, we determined that STAT3 activation occurs in response to G-CSF but not in response to ATRA. Neither short-term (15 and 90 minutes, not shown) nor longer term (24-72 hours, Figure 5, lanes 2-4) ATRA induction induced phosphorylation of STAT3. Our studies in EPRO-Gr cells suggest that G-CSF–induced signals can overcome the promyelocyte differentiation arrest associated with the dominant negative RARα receptor. Accordingly, we investigated whether the lack of G-CSF responsiveness in APL cells could be attributed to the absence of G-CSFR expression. Both Northern blot analysis (Figure 4A) and Western blotting (Figure 5) confirm that G-CSFR is not expressed in uninduced NB4 and is up-regulated in response to ATRA. We therefore examined STAT3 phosphorylation in NB4 cells before and after ATRA induction. Uninduced and ATRA-treated NB4 cells were exposed to a 15-minute G-CSF pulse and lysates assayed for STAT3 phosphorylation. STAT3 phosphorylation was evident only after a G-CSF pulse once the cells were induced to express G-CSFR with ATRA (Figure 5, lanes 5-8), suggesting that ATRA induces functional G-CSFR expression. Total STAT3 protein levels were not sensitive to ATRA or G-CSF induction and remained unchanged. (Figure 5, bottom panel). These observations suggest that although ATRA induces expression of G-CSFR in NB4 cells, STAT3 phosphorylation depends on G-CSFR signaling, thus offering a possible basis for the previously observed synergy between ATRA and G-CSF in mediating APL cell differentiation.
STAT1 and STAT5 phosphorylation was not detected in ATRA-induced or G-CSF–primed ATRA-induced NB4 cells (data not shown).
Effect of G-CSFR activation on morphologic differentiation and neutrophil gene expression in NB4 cells
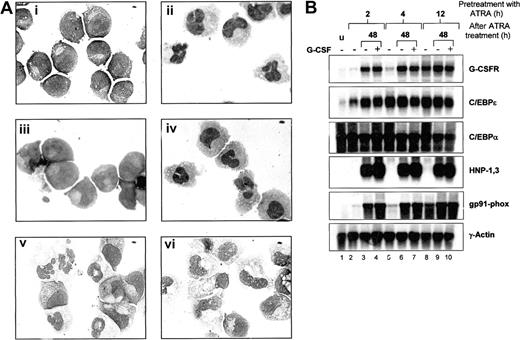
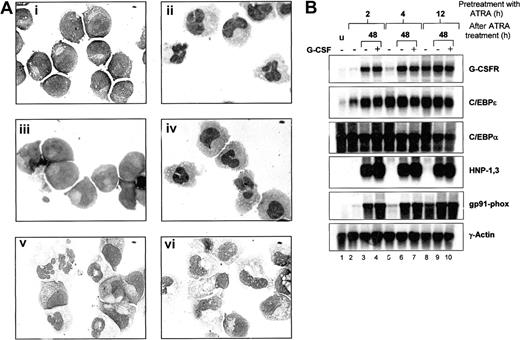
We next investigated whether ATRA-induced G-CSF responsiveness could enhance morphologic maturation of NB4 cells. NB4 cells were continuously exposed to ATRA, G-CSF, or ATRA plus G-CSF and examined for morphologic differentiation. As previously observed, NB4 cells treated with G-CSF alone did not undergo morphologic differentiation (Figure 6A and Khanna-Gupta et al34 ). NB4 cells exposed to ATRA plus G-CSF were indistinguishable from cells differentiated with ATRA alone (Figure 6A). We next tested whether short-term exposure of NB4 cells to ATRA could “prime” cells by inducing G-CSFR expression, such that subsequent induction with G-CSF enhanced maturation. NB4 cells were pretreated with ATRA for 2, 4, 6, 8, 12, and 24 hours, then washed and induced either in maintenance media (without ATRA) or in maintenance media with added G-CSF. At 24-hour intervals, cells were examined for morphology, and RNA was extracted for northern analysis of neutrophil gene expression. NB4 cells primed with ATRA for only 2 hours and grown in maintenance media alone showed very little change in morphology, whereas cells primed for 4 to 12 hours showed increasing, but modest (at 12 hours, < 30% of cells), levels of morphologic maturation over 3-4 days. Cells primed with ATRA for 24 hours and then grown in maintenance media demonstrated complete morphologic maturation after 72 hours (Figure 6A). The inclusion of G-CSF in the maintenance media following ATRA priming had little effect on differentiation.
Effects of G-CSFR activation on ATRA-induced morphologic maturation and neutrophil gene expression in NB4 cells. (A) NB4 cells (i) were induced with 5 μM ATRA alone (ii), G-CSF alone (iii), ATRA plus 100ng/mL G-CSF (iv), or were primed with ATRA for 24 hours, washed, and then incubated in ATRA-free media either in the absence (v) or the presence of G-CSF (vi). Following 4 days of incubation, cells then were cytocentrifuged and stained with Wright-Geimsa stain for morphologic analysis. Original magnification, × 100. (B) Aliquots of NB4 cells (u) primed with ATRA (5 μM) for 2, 4, and 12 hours were either extracted for total RNA (lanes 2, 5, and 8) or washed in 1 × PBS and incubated for 48 hours in ATRA-free maintenance media alone (lanes 3, 6, and 9) or in the presence of G-CSF (100 ng/mL) (lanes 4, 7, and 10). After the 48-hour incubation period, cells in media alone (–) or in media with G-CSF (+) were collected and subjected to Wright-Geimsa staining or extracted for RNA. Total RNA (10 μg) from each sample was then blotted and sequentially hybridized with 32P-labeled cDNA probes for human G-CSFR, C/EBPϵ, HNP-1,3 and gp91-phox, and rat C/EBPα. The blot was also hybridized with human γ-actin to demonstrate equal loading of RNA in each lane.
Effects of G-CSFR activation on ATRA-induced morphologic maturation and neutrophil gene expression in NB4 cells. (A) NB4 cells (i) were induced with 5 μM ATRA alone (ii), G-CSF alone (iii), ATRA plus 100ng/mL G-CSF (iv), or were primed with ATRA for 24 hours, washed, and then incubated in ATRA-free media either in the absence (v) or the presence of G-CSF (vi). Following 4 days of incubation, cells then were cytocentrifuged and stained with Wright-Geimsa stain for morphologic analysis. Original magnification, × 100. (B) Aliquots of NB4 cells (u) primed with ATRA (5 μM) for 2, 4, and 12 hours were either extracted for total RNA (lanes 2, 5, and 8) or washed in 1 × PBS and incubated for 48 hours in ATRA-free maintenance media alone (lanes 3, 6, and 9) or in the presence of G-CSF (100 ng/mL) (lanes 4, 7, and 10). After the 48-hour incubation period, cells in media alone (–) or in media with G-CSF (+) were collected and subjected to Wright-Geimsa staining or extracted for RNA. Total RNA (10 μg) from each sample was then blotted and sequentially hybridized with 32P-labeled cDNA probes for human G-CSFR, C/EBPϵ, HNP-1,3 and gp91-phox, and rat C/EBPα. The blot was also hybridized with human γ-actin to demonstrate equal loading of RNA in each lane.
We next examined neutrophil-specific gene expression following ATRA priming in the absence or presence of G-CSF. Consistent with our previous observations, up-regulation of G-CSFR expression required at least 4 hours of ATRA treatment (Figure 6B, lanes 2, 5, and 8). Increased expression of C/EBPϵ, on the other hand, appeared by 2 hours of induction, with maximal levels at 4 hours of induction (Figure 6B, C/EBPϵ panel, lanes 2 and 5). C/EPBα expression remained high throughout the induction process. Interestingly, expression of G-CSFR continued to increase in NB4 cells primed with ATRA for only 2 hours, with subsequent incubation in maintenance media for 48 hours either in the absence or presence of G-CSF (Figure 6B, G-CSFR panel, lanes 3 vs. 4, 6 vs. 7, and 9 vs. 10). C/EBPϵ expression also remained high, despite removal of ATRA from the medium. Furthermore, expression of 2 neutrophil markers, the human α defensins HNP-1,3 and the myeloid cytochrome heavy chain gene gp91-phox, also markedly increased in 2 hours' ATRA-primed NB4 cells incubated in ATRA-free maintenance media for 48 hours, again either in the absence or presence of G-CSF. Importantly, all of the observed changes in gene expression in NB4 cells following 2 hours of ATRA priming occurred in the absence of morphologic maturation, and the inclusion of G-CSF in the media did not affect either morphologic differentiation or gene expression. No induction regimen up-regulated lactoferrin expression (data not shown).
Discussion
The addition of G-CSF during ATRA-mediated differentiation of APL cells has been reported to augment neutrophil differentiation over that seen with ATRA alone.10-12 Additionally, ATRA-resistant APL can be induced to differentiate by the combination of G-CSF and ATRA.13,14 To elucidate the mechanism underlying these observations, we developed a model to separate the effects of RA and G-CSF signaling during neutrophil maturation. We used the EML/EPRO cell line in which a dominant negative RARα leads to a myeloid differentiation arrest.21 We rendered EPRO cells G-CSF responsive by generating EPRO-Gr cells overexpressing the G-CSFR. Unlike parental EPRO cells or EPRO cells transduced with vector alone, EPRO-Gr cells expressed high levels of G-CSFR, and upon induction with G-CSF underwent morphologic and biochemical differentiation to segmented neutrophils. These results indicate that EPRO cells express the necessary signaling mechanisms for G-CSF–mediated differentiation, and when G-CSFR levels are sufficiently high, the cells become fully G-CSF responsive. Furthermore, G-CSF–induced EPRO-Gr differentiation occurred without ATRA despite the presence of a dominant-negative retinoic acid receptor. G-CSF–induced differentiation of the EPRO-Gr cells also occurred without concomitant increases in C/EBPϵ expression, whereas the presence of ATRA significantly increased C/EBPϵ expression. This result is consistent with previous studies demonstrating C/EBPϵ is a direct retinoid target.18 However, whether C/EBPϵ expression is important to G-CSF–induced signaling pathways is unclear, since there appears to be significant levels of C/EBPϵ expression in both uninduced and G-CSF–induced EPRO-Gr cells.
The mechanism by which ATRA induces myeloid differentiation in APL is unknown. Proponents of an active role suggest that ATRA functions through RAREs to activate transcription, initiating a cascade of genetic events leading to differentiation. Alternatively, it has been hypothesized that dominant-negative forms of RARα found in APL interfere with other trans-acting elements necessary for the normal neutrophil differentiation program.9 An additional hypothesis is that RA target genes can be transactivated by ATRA-independent mechanisms: IL-3–mediated myeloid differentiation of EML cells has been reported to be associated with ATRA-independent activation of RAREs.38 To examine this question, we evaluated the expression of a RARE-containing reporter plasmid in EPRO-Gr cells. As expected, ATRA-mediated differentiation was associated with transactivation of a RARE-luciferase reporter plasmid. In contrast, G-CSF–mediated differentiation of EPRO-Gr cells was not associated with RARE-mediated transactivation. Unlike the IL-3–induced transactivation of a RARE containing reporter plasmid in EML cells reported by Johnson et al,38 induction of EPRO-Gr with G-CSF was not associated with ATRA-independent RARE activation. Therefore, G-CSF presumably induces neutrophil differentiation of EPRO-Gr cells by a RARE independent pathway.
G-CSF mediates neutrophil differentiation via G-CSFR activation with subsequent activation of the JAK2/STAT3 signal transduction pathway3 associated with tyrosine phosphorylation of STAT3 by JAK2.4,33 We therefore determined whether differentiation of EPRO-Gr was associated with phosphorylation of STAT3. Uninduced and ATRA-induced cells expressed only unphosphorylated STAT3 protein, while G-CSF induction resulted in STAT3 phosphorylation. These findings suggest that ATRA-mediated differentiation of EPRO-Gr occurs independent of the STAT3 activation associated with G-CSF–induced maturation. Although STAT1 and STAT5 also have been implicated in G-CSF–mediated myeloid differentiation, we saw no modulation of STAT1 or STAT5 phosphorylation with G-CSF or ATRA induction of EPRO-Gr.
In EPRO-Gr cells, ATRA apparently mediates differentiation via activation of RA-target genes, while G-CSF induces differentiation via activation of STAT3 target genes. These results raise the question as to how such divergent pathways can elicit similar responses, and therefore the interaction of these pathways was examined in APL cells. Previous reports have suggested that G-CSF can augment the extent of neutrophil differentiation of APL cells induced with ATRA.10-12 Additionally, ATRA-resistant APL cells have been reported to differentiate with the combination of G-CSF and ATRA.13-15 It is noteworthy that G-CSF alone does not lead to neutrophil differentiation of APL.34
Uninduced NB4 cells expressed little or no G-CSFR, and ATRA induction up-regulated G-CSFR mRNA and protein levels within 24 hours. This up-regulated G-CSFR expression induced by ATRA appeared to require protein synthesis: expression of G-CSFR required more than 4 hours of ATRA induction and was blocked when induction was performed in the presence of cycloheximide. Therefore, G-CSFR is not a direct target of ATRA. Since regulators of G-CSFR expression in myeloid cells include members of C/EBP family of transcription factors, in particular, C/EBPα,27 we examined C/EBP mRNA levels in NB4 cells. While C/EBPα expression was constitutive and not modulated by ATRA, C/EBPϵ expression was up-regulated by ATRA in an immediate-early fashion, with increased expression at 2 hours even in the presence of cycloheximide. This is consistent with previous reports that C/EBPϵ is a direct target of RA signaling.39 Hence, G-CSFR is unexpressed in NB4 cells despite adequate levels of its putative regulator C/EBPα. Furthermore, its transcriptional activation may, in part, be regulated by C/EBPϵ. Our ChIP data demonstrate that both C/EBPα and C/EBPϵ bind the G-CSFR promoter in uninduced NB4 cells. While C/EBPϵ remains bound to the G-CSFR promoter, C/EBPα binding appears to be reduced in ATRA-induced NB4 cells. This suggests that decreased C/EBPα binding to the G-CSFR promoter may actually contribute to the ATRA-induced expression of G-CSFR in NB4 cells, perhaps by allowing formation of C/EBPϵ homodimers or of C/EBPϵ heterodimers with other binding partners. This further suggests that the proposed C/EBPα sequestration by the PML-RARα gene product in APL cells does not explain the lack of G-CSFR expression in uninduced NB4 cells.
Despite the up-regulation of G-CSFR, NB4 cells, like EPRO cells, appear to differentiate in ATRA via a G-CSF/STAT3-independent pathway. We hypothesized that G-CSFR expression induced by ATRA, however, should render NB4 cells G-CSF responsive. As expected, NB4 cells induced with ATRA phosphorylate STAT3 in response to G-CSF. These findings suggest that, like EPRO-Gr cells, NB4 cells are capable of mediating both RA and G-CSF–mediated signals. Thus, the ability of G-CSF to augment ATRA-mediated differentiation, as well as its ability to differentiate ATRA resistant APL cells, may reflect the ability of ATRA to up-regulate G-CSFR expression. This may also explain the absence of an in vivo and in vitro effect by G-CSF alone on APL cells. However, we were unable to demonstrate an association between G-CSFR signaling through STAT3 and enhanced morphologic maturation. Pretreatment of NB4 cells with ATRA for less than 12 hours, with subsequent transfer to media, induced only modest morphologic maturation that were not enhanced by G-CSF. Conversely, pretreatment of NB4 cells with ATRA for 24 hours led to terminal maturation even upon removal of ATRA and did not require any growth factor. Finally, short-term (2 hours) priming with ATRA caused up-regulation of C/EBPϵ and the 2 neutrophil genes HNP 1, 3, and gp91-phox that persisted in the absence of ATRA or G-CSF. These results indicate that during ATRA induction of NB4 cells, 2 thresholds are reached. The first is reached within 2 hours of ATRA induction, beyond which neutrophil gene expression increases autonomously. The second threshold is reached after 12 hours of exposure to ATRA, whereupon the mechanisms that drive terminal morphologic differentiation proceed to completion. Neither of these steps is enhanced by G-CSF, despite intact STAT3-mediated signaling. These results suggest that the synergistic effect of G-CSF and ATRA on maturation of t(15;17) APL cells or on overcoming ATRA resistance in t(15;17) and t(11;17) APL cells does not simply reflect increased G-CSFR expression providing G-CSF responsiveness. It may be influenced by the leukemic phenotype induced by the leukemogenic fusion proteins, which function as more than simple dominant-negative RARs. To further address this issue, cells expressing ATRA-resistant variants of RARα fusion proteins and primary APL cells must be examined for effects of G-CSF on ATRA-induced differentiation. We are currently pursuing such studies, which may help to elucidate the molecular mechanisms underlying the ability of G-CSF to augment ATRA-induced granulocytic differentiation and may lead to novel therapeutic approaches to APL.
Prepublished online as Blood First Edition Paper, November 6, 2003; DOI 10.1182/blood-2002-10-3247.
Supported by National Institutes of Health awards R01-DK53471 and P01-HL63357 (N.B.), F32-DK09744 (N.A.M.), and K01-DK60565 (P.G.).
N.A.M. and P.G. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.