Abstract
We report here 7 new mutations in the ADAMTS13 gene responsible for Upshaw-Schulman syndrome (USS), a catastrophic phenotype of congenital thrombotic thrombocytopenic purpura, by analyzing 5 Japanese families. There were 3 mutations that occurred at exon-intron boundaries: 414+1G>A at intron 4, 686+1G>A at intron 6, and 1244+2T>G at intron 10 (numbered from the A of the initiation Met codon), and we confirmed that 2 of these mutations produced aberrantly spliced messenger RNAs (mRNAs). The remaining 4 mutations were missense mutations: R193W, I673F, C908Y, and R1123C. In expression experiments using HeLa cells, all mutants showed no or a marginal secretion of ADAMTS13. Taken together with the findings in our recent report we determined the responsible mutations in a total of 7 Japanese patients with USS with a uniform clinical picture of severe neonatal hyperbilirubinemia, and in their family members, based on ADAMTS13 gene analysis. Of these patients, 2 were homozygotes and 5 were compound heterozygotes. The parents of one homozygote were related (cousins), while those of the other were not. Molecular models of the metalloprotease, fifth domain of thrombospondin 1 (Tsp1-5), and Tsp1-8 domains of ADAMTS13 suggest that the missense mutations could cause structural defects in the mutants.
Introduction
Thrombotic thrombocytopenic purpura (TTP) is a life-threatening generalized disorder, and its diagnosis is made according to the criteria of Moschcowitz's pentad1 : thrombocytopenia, microangiopathic hemolytic anemia (MAHA), fluctuating neurologic signs, renal failure, and fever. These criteria, however, are almost undistinguishable from those of hemolytic-uremic syndrome (HUS) with Gasser's triad2 ; MAHA, thrombocytopenia, and renal insufficiency. Thus, the comprehensive term “TTP/HUS” or “thrombotic microangiopathy”3 has frequently been used in clinical practice.
Recent advances in elucidating the proteolytic processing of plasma von Willebrand factor (VWF) multimers have established assays for the activity of VWF-cleaving protease and its inhibitor (autoantibody).4-7 These assays have largely made it possible to distinguish TTP from HUS, because the former has defective VWF-cleaving activity, whereas the latter has VWF-cleaving activity.6,7 Studies by several groups of investigators have led to the identification of this enzyme as a new metalloprotease belonging to the ADAMTS (a disintegrinlike and metalloprotease with thrombospondin type 1 motif) family, which has been designated ADAMTS13.8-12 This enzyme is produced in the liver.10-12 The deduced amino acid residue number is 1427, and the gene contains 29 exons and is located on chromosome 9q34.10-12
Upshaw-Schulman syndrome (USS) was originally reported as a disease complex with repeated episodes of thrombocytopenia and hemolytic anemia that quickly respond to infusions of fresh frozen plasma (FFP).13-16 Clinical signs often develop in the patients during the newborn period or early infancy. In fact, the earliest and most frequently encountered clinical manifestation is severe hyperbilirubinemia with negative Coombs test soon after birth, which requires exchange blood transfusions. Pediatric hematologists have long been more familiar with this disease than general physicians, but this diagnostic name appears to be lacking or ignored in most medical textbooks because of its uncertain clinical entity. Thus, a variety of alternative nomenclatures has been given to this disease, such as chronic relapsing TTP, congenital MAHA, and familial TTP/HUS.
Under these circumstances, the report of Furlan et al17 in 1997 showing that 4 cases of chronic relapsing TTP lacked ADAMTS13 activity was notable. Of these cases, 2 were siblings with a consistent deficiency of the enzyme activity, suggesting that they had an inheritable form, whereas the other 2 were unrelated and apparently had an acquired form. Since then, there have been several reports on TTP/HUS associated with congenital deficiency of ADAMTS13 activity, and, accordingly, the diagnostic name of USS has been re-evaluated and has gained a position as an independent disease entity. Usually, parents of USS patients have reduced plasma ADAMTS13 activity, roughly half of the normal control level, indicating that they are asymptomatic carriers.15 Among them, one father detected in our laboratory was particularly interesting because he had an extremely low level of plasma ADAMTS13 activity, 4.5% to 7% of the normal control level on 3 different occasions, but so far, at the age of 36, he has no apparent clinical signs.15 The unusually low level of ADAMTS13 activity in this man was further confirmed by both genetic and biochemical analyses,18 that is, he was compound heterozygote, R268P/P475S. In expression analysis, the R268P mutant was not secreted from the cells, but the P475S mutant was secreted normally and showed low but significant activity. The P475S mutation is prevalent in the Japanese population (heterozygosity ∼ 10%). Thus, the discovery of new ADAMTS13 mutations in different countries or races appears to be very important for the analysis of a potential linkage with other thrombotic generic risks.
Here, we identified 7 new mutations in the ADAMTS13 gene responsible for USS by analyzing 5 patients and 16 relatives belonging to 5 different families from widely separated regions in Japan. All the patients had an episode of severe hyperbilirubinemia during the newborn period, and received exchange blood transfusions, except for one. Furthermore, structural changes of the mutant ADAMTS13 molecules associated with impaired enzyme activity were predicted using a homology modeling method.
Patients, materials, and methods
Families A to G with USS
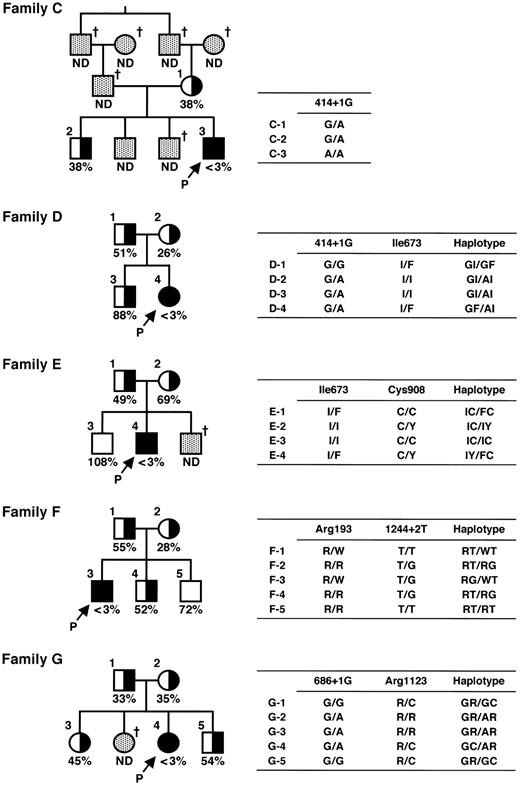
Families C to G are shown in Figure 1.
Pedigree and haplotypes of patient families. Squares and circles indicate males and females, respectively. Closed circles and squares with arrows with a P indicate probands. The half-closed circles and squares represent asymptomatic carriers. The cross indicates deceased. The ADAMTS13 activity is shown as a percentage of the normal control. ND indicates not determined. Mutations found in the ADAMTS13 gene are shown as one-letter amino acid abbreviations numbered from the initial Met or as nucleotides numbered from the A of the translation initiation Met codon.
Pedigree and haplotypes of patient families. Squares and circles indicate males and females, respectively. Closed circles and squares with arrows with a P indicate probands. The half-closed circles and squares represent asymptomatic carriers. The cross indicates deceased. The ADAMTS13 activity is shown as a percentage of the normal control. ND indicates not determined. Mutations found in the ADAMTS13 gene are shown as one-letter amino acid abbreviations numbered from the initial Met or as nucleotides numbered from the A of the translation initiation Met codon.
Families A and B. There were 2 USS probands, A and B, belonging to different families described and characterized in detail in a recent publication.18
Family C. Proband C is a male born in Fukui in 1972 whose history during childhood was reported in 1984.19 His parents are cousins. His father died of cerebral infarction at 63 years of age, and the third brother died of melena soon after birth. His mother and 2 other brothers have no thrombotic or hemorrhagic signs. Proband C showed hyperbilirubinemia during the newborn period, and he received phototherapy for 3 days without exchange blood transfusion. Thereafter, he had repeated episodes of thrombocytopenia and hemolytic anemia, and he has received prophylactic infusion of 2 units (160 mL) of FFP every 2 to 4 weeks since he was 8 years old. However, he has gradually developed chronic nephritis and was required to receive continuous ambulatory peritoneal dialysis starting in March 1995. Because of repeated peritonitis associated with continuous ambulatory peritoneal dialysis, however, his therapy for renal insufficiency was completely switched to hemodialysis starting in May 1999.
Family D. Proband D is a female born in Yamaguchi in 1978 whose history until the age of 4 years was reported in 1982.20 Her parents are unrelated. She had an episode of severe hyperbilirubinemia soon after birth that required 2 exchange blood transfusions. Since 4 years of age, she received 1 unit (80 mL) of FFP every 3 weeks until the age of 21 years. Under this treatment regimen, however, her renal function test, including serum creatinine level, was getting worse, and therefore the volume of FFP infused was increased stepwise. She now receives 5 units (400 mL) of FFP infusion every 2 weeks for prophylaxis.
Family E. Proband E is a male born in Kagawa in 1985. He is the second child of unrelated parents. His younger brother with Down syndrome died of an unknown cause soon after birth. His parents and elder brother are apparently healthy. This proband developed severe hyperbilirubinemia and thrombocytopenia in the next day after birth. He received 2 exchange blood transfusions on the second day of life, resulting in excellent clinical improvement. At 5 years of age, he had an episode of thrombocytopenia and hemolytic anemia. Such clinical manifestations quickly improved after 2 plasma exchanges, together with the correction of abnormal laboratory test results that confirmed a clinical diagnosis of USS. After that, he receives 2 to 3 units (160-240 mL) of FFP infusion upon episodes of thrombocytopenia and hemolytic anemia.
Family F. Proband F is a male born in Aomori in 1993. He is the first child of unrelated parents. His parents and brothers are all apparently healthy. Soon after birth, this proband developed severe hyperbilirubinemia and received 2 exchange blood transfusions. At 10 months after birth, he had generalized petechiae with hemolytic anemia and thrombocytopenia. Therefore, he was once diagnosed with idiopathic thrombocytopenic purpura. At 2½ years of age, he received 1 unit (80 mL) of FFP infusion for the aforementioned clinical signs, which dramatically improved both his clinical and laboratory findings, resulting in a clinical diagnosis of USS. Now, he receives 1 unit of FFP infusion for each of his occasional hemolytic crises.
Family G. Proband G is a female born in Tokyo in 1987. She is the third child of unrelated parents. One elder sister had an episode of severe hyperbilirubinemia soon after birth and received exchange blood transfusion. She died of intracranial bleeding after a traffic accident at 8 years old. Her parents, remaining elder sister, and brother are all apparently healthy. Soon after birth the proband developed severe hyperbilirubinemia and received exchange blood transfusion. Thereafter, she had repeated episodes of thrombocytopenia and hemolytic anemia that were quickly improved by FFP infusion. Finally, at 14 years of age, she was clinically diagnosed with USS. Now, she receives 10 mL/kg FFP infusion for each of her occasional hemolytic crises.
Assays of ADAMTS13 activity
Plasma ADAMTS13 activity was assayed by the method of Furlan et al4,6 based on VWF multimer analysis, with a slight modification as described.15 The ADAMTS13 activity of pooled normal plasma was defined as 100%. The normal range of ADAMTS13 activity (n = 60; 30 women and 30 men, 20-39 years of age) was 102 ± 23% (mean ± 1 SD).21
Sequencing of the ADAMTS13 gene
All DNA experiments were performed with the permission of the ethics committees of both the sample-collecting hospital and the gene-analyzing institute. Amplification and sequencing of the 29 exons of the ADAMTS13 gene were performed as recently described.18
Transient expression of ADAMTS13
Polymerase chain reaction (PCR)–based mutagenesis was performed for the construction of ADAMTS13 mutants as recently reported.18 The mutant cDNA was cloned into mammalian expression vector, pCAGG.22 The DNA sequence of all inserts was confirmed by DNA sequencing.
Each of the expression vectors was transfected into HeLa cells using FuGENE6 (Roche Molecular Biochemicals, Indianapolis, IN), according to the manufacturer's instructions. Briefly, HeLa cells were cultured in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen) in humidified air with 5% CO2 at 37°C. Of each expression plasmid, 5 μg was transfected into subconfluent cells in 90-mm dishes. After 4 to 6 hours of incubation, the medium was changed to 4 mL serum-free OPTI-MEM I (Invitrogen), and the cultures were incubated for 44 hours. The media were collected and the culture media were concentrated to one tenth the original volume. The cells were washed with phosphate-buffered saline, pH 7.4, and lysed with 300 μL sodium dodecyl sulfate (SDS) sample buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]–HCl/2% SDS/50 mM dithiothreitol/10 mM ethylenediaminetetraacetic acid/0.02% bromophenol blue/6% glycerol, pH 6.8).
Western blot analysis
The media and cell lysates were first separated by SDS–polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane (BioRad, Hercules, CA). After blocking with 3% skim milk, the membrane was incubated with 1 μg/mL anti-FLAG (fludarabine, cytarabine, and granulocyte colony-stimulating factor) M2 monoclonal antibody (Sigma, St Louis, MO) and then 0.1 μg/mL peroxidase-labeled goat antimouse immunoglobulin G (Kirkegaard & Perry Laboratories, Gaithersburg, MD). Luminographic detection was performed using the Western Lighting Chemiluminescence Reagent (PerkinElmer Life Sciences, Shelton, CT) and detected using an image analyzer LAS 1000 plus (Fujifilm, Tokyo, Japan).
Reverse-transcription PCR
To evaluate the transcription products of the gene with a single nucleotide mutation in introns 4 and 10, we performed reverse transcription (RT)–PCR on mRNA using primers corresponding to the respective exons. RT-PCR for intron 4 using sense primer GGGCAGAACTGCTTCGGGACC (exon 4) and antisense primer AGCATGGCCAGGATCCGTGTC (exon 5) yields a 185–base pair (bp) band from the normally spliced transcript or a 510-bp band from the unspliced transcript. RT-PCR for intron 10 using sense primer GGGTCCCCGAAGTCCTTGCTC (exon 10) and antisense primerAGGTCAGCACCAACACATGCA (exon 11) yields a 115-bp band from the spliced transcript or a 226-bp band from the unspliced transcript. The mRNAs were prepared using a PAXgene Blood RNA system (Qiagen, Valencia, CA), and RT-PCR was performed using Qiagen OneStep RT-PCR (Qiagen).
Construction of 3-dimensional models of metalloprotease, Tsp1-5, and Tsp1-8 domains of ADAMTS13
Model structures of the wild-type (WT) metalloprotease domain, and fifth and eighth domains of thrombospondin 1 (Tsp1-5 and Tsp1-8 domains) were constructed based on homology modeling methods. Searching for the reference proteins and sequence alignments was performed using position-specific iterated (PSI)–BLAST.23 The reference proteins were adamalysin II (PDB24 ID: 4AIG25 ) for the metalloprotease domain and thrombospondin-1 type 1 repeats (PDB ID: 1LSL26 ) for Tsp1-5 and Tsp1-8 domains. The sequence alignments produced using PSI-BLAST were manually adjusted taking biologically important regions and secondary structure into consideration using the CHIMERA modeling system.27,28 The model structures were constructed using the fully automatic modeling system FAMS29,30 based on each alignment.
Results
ADAMTS13 activity and ADAMTS13 inhibitor in patients with USS and their families
The family pedigrees of 5 patients and their ADAMTS13 activity are shown in Figure 1. The plasma ADAMTS13 activities of all probands were less than 3% of the normal control value. The low activity (<3%) of ADAMTS13 in the proband was confirmed using the plasmas of at least 2 different occasions, with an interval of more than 6 months. The ADAMTS13 activity, which was less than 3% of the normal, of 2 other USS patients A and B in 2 different families was described in our previous paper.17 Neither the patients' nor their relatives' plasmas contained detectable inhibitor of ADAMTS13 activity (<0.5 Bethesda unit/mL).
The earliest uniform clinical picture of these 7 patients was severe-to-moderate hyperbilirubinemia during the newborn period that required exchange blood transfusion, except in one case (patient C), who was treated with phototherapy. After the clinical diagnosis of USS was made, all patients were treated with a prophylactic infusion of 5 to 10 mL of FFP/kg at 2- to 3-week intervals or with FFP infusion when they had clinical manifestations.
ADAMTS13gene mutations in patients with USS and their families
We analyzed the ADAMTS13 gene in 5 patients with USS and 21 of their family members by PCR amplification and sequencing of the 29 exons of the gene. Single-nucleotide mutations at 12 sites were identified (Table 1). There were 4 silent mutations identified: 420T>C, 1716G>A, 2280C>T, and 4221C>A, and all of these mutations have been reported as single nucleotide polymorphisms (SNPs).12,18 We identified 8 additional mutations in the 5 families. Of 8 mutations, 5 found in the exons were missense mutations: 577C>T (R193W), 1342C>G (Q448E), 2017A>T (I673F), 2723G>A (C908Y), and 3367C>T (R1123C). Of these, 1342C>G (Q448E) has been reported as an SNP.12,18 The remaining 3 mutations were found in introns at +1 or +2 nucleotides from the exon-intron boundary, and these mutations in introns appeared to render the gene unable to produce a normally spliced transcript.
To investigate the frequencies of the 7 mutations newly identified in this report, we performed sequencing of exons 4, 6, 10, 17, 21, and 25 of the ADAMTS13 gene in genomic DNAs isolated from 96 Japanese individuals without TTP. They were all patients of the Division of Hypertension and Nephrology at the National Cardiovascular Center (Suita, Japan). None of these 7 mutations appeared in this panel, indicating that they were rare (<1% heterozygosity) in the Japanese population.
Expression of rADAMTS13 mutants
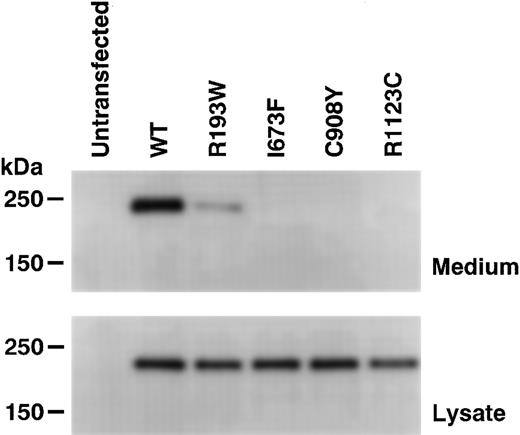
To determine the effects of the newly identified mutations, WT and mutant forms of recombinant ADAMTS13 (rADAMTS13) were transiently expressed in HeLa cells. The expression of WT rADAMTS13 produced a single immunoreactive band with a molecular mass of about 230 kDa in the culture medium (Figure 2, top). The band was not detected in the medium of untransfected cells. In the culture medium of cells expressing the R193W mutant, a weak band was detected with the same size as WT. The expression level of the R193W mutant was roughly estimated as one fourth of WT by comparing their chemiluminescent intensities on Western blot. On the other hand, no bands were detected for the I673F, C908Y, and R1123C mutants, although these mutants were synthesized within the cells (Figure 2, bottom). These results clearly indicated that these mutants are not secreted normally.
Expression of rADAMTS13. The wild-type (WT) rADAMTS13 and mutants with C-terminal FLAG-tag were transiently expressed in HeLa cells. The culture media (top) and cell lysates (bottom) were analyzed by Western blotting with an anti-FLAG antibody. The sizes of protein standards are indicated at the left.
Expression of rADAMTS13. The wild-type (WT) rADAMTS13 and mutants with C-terminal FLAG-tag were transiently expressed in HeLa cells. The culture media (top) and cell lysates (bottom) were analyzed by Western blotting with an anti-FLAG antibody. The sizes of protein standards are indicated at the left.
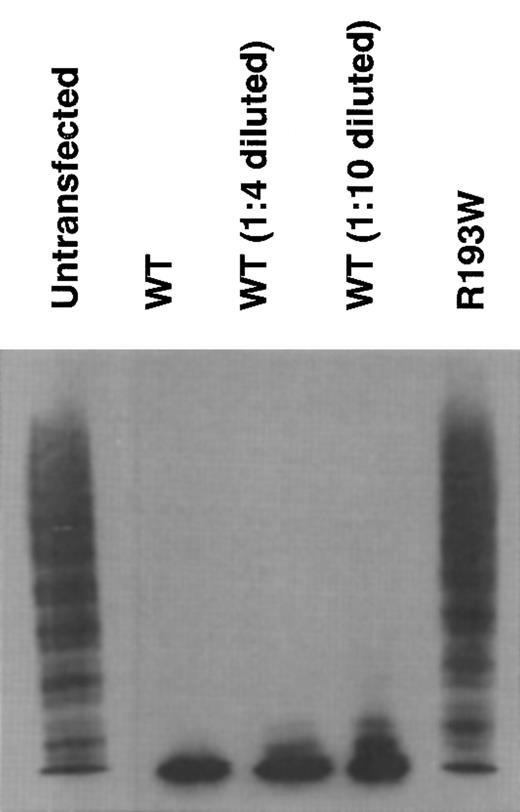
The enzymatic activity of rADAMTS13 was determined by analyzing the degradation of VWF multimers.18 In the medium of untransfected cells, the ladders of VWF multimers extended into the high-molecular-weight area (Figure 3, untransfected), indicating the lack of ADAMTS13 activity in the medium. In contrast, the ladder diminished after incubation with the medium of transfected cells expressing WT rADAMTS13, indicating that WT has ADAMTS13 activity. This activity was also observed in the 1:4- and 1:10-diluted WT. As for the R193W mutant, no enzymatic activity was observed (Figure 3), indicating that the R193W mutant had little activity.
Cleavage of VWF multimer by rADAMTS13. The rADAMTS13 activity was measured by degradation of VWF multimers. The culture media of untransfected, WT, and the R193W mutant were incubated with purified VWF. The multimeric state of VWF was determined by Western blot analysis after SDS–agarose gel electrophoresis. The 1:4 diluted WT medium contained the enzyme amount roughly equivalent to the medium of the R193W mutant.
Cleavage of VWF multimer by rADAMTS13. The rADAMTS13 activity was measured by degradation of VWF multimers. The culture media of untransfected, WT, and the R193W mutant were incubated with purified VWF. The multimeric state of VWF was determined by Western blot analysis after SDS–agarose gel electrophoresis. The 1:4 diluted WT medium contained the enzyme amount roughly equivalent to the medium of the R193W mutant.
Aberrant splicing caused by mutations at exon-intron boundary
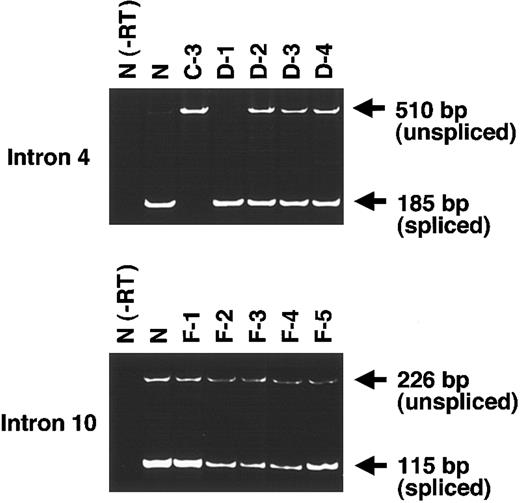
To test the effects of the mutations in introns 4 and 10, we performed RT-PCR using exon primers. Since no bands corresponding to the RT-PCR products were detected without the reverse-transcription reaction in normal control samples, the possibility that bands were generated due to the contaminating genomic DNA was excluded (Figure 4, lane N(-RT)).
Effects of intronic mutations on mRNA splicing. To evaluate the products of genes with a single splice mutation in intron 4 (top) or intron 10 (bottom), we performed RT-PCR on mRNA using primers corresponding to these exons. RT-PCR for intron 4 yielded a 185-bp band from the normally spliced transcript and a 510-bp band from the unspliced transcript. RT-PCR for intron 10 yielded a 115-bp band from the spliced transcript and a 226-bp band from the unspliced transcript.
Effects of intronic mutations on mRNA splicing. To evaluate the products of genes with a single splice mutation in intron 4 (top) or intron 10 (bottom), we performed RT-PCR on mRNA using primers corresponding to these exons. RT-PCR for intron 4 yielded a 185-bp band from the normally spliced transcript and a 510-bp band from the unspliced transcript. RT-PCR for intron 10 yielded a 115-bp band from the spliced transcript and a 226-bp band from the unspliced transcript.
For the 414+1G>Amutation in intron 4, we examined mRNAfrom 6 individuals: healthy control (N), C-3 (homozygote for the mutation), D-1 (no mutation), and D-2 to D-4 (heterozygotes) (Figure 4, top). In N and D-1, only one band of 185 bp corresponding to the normally spliced product was detected. In D-2 to D-4, a 510-bp band corresponding to unspliced transcript was found together with the 185-bp band derived from the spliced transcript. In contrast, in C-3, only one band of 510 bp was found. Thus, it was clear that the mutation 414+1G>A abolished the splicing at the exon 4–intron 4 boundary.
For the 1244+2T>G mutation in intron 10, we similarly performed RT-PCR on mRNA from 6 individuals: N, F-1 and F-5 (no mutation), and F-2 to F-4 (heterozygotes) (Figure 4, bottom). In this experiment, we were unable to obtain a sample of a homozygote for the mutation because the proband with this mutation was compound heterozygote. As a result, the product from all 6 individuals had 2 bands of 115 bp derived from the spliced transcript and 226 bp derived from the unspliced transcript. However, it is clear that the intensity of the 115-bp band of F-1 and F-5 was much denser than that of the 226-bp band. In contrast, the intensity of the 115-bp band of F-2 to F-4 was much weaker than that of N, F-1, and F-5. Based on these results, intron 10 appears to be not completely spliced even in healthy individuals, suggesting that in healthy individuals alternative splicing occurred at exon 10–intron 10 boundary or the splicing of this boundary had low efficiency. Furthermore, the transcripts of heterozygous individuals are likely to be less spliced than those of healthy individuals. Thus, the mutation of 1244+2T>G abolishes splicing at the exon-intron boundary.
Molecular modeling of the metalloprotease and Tsp1 domains of ADAMTS13
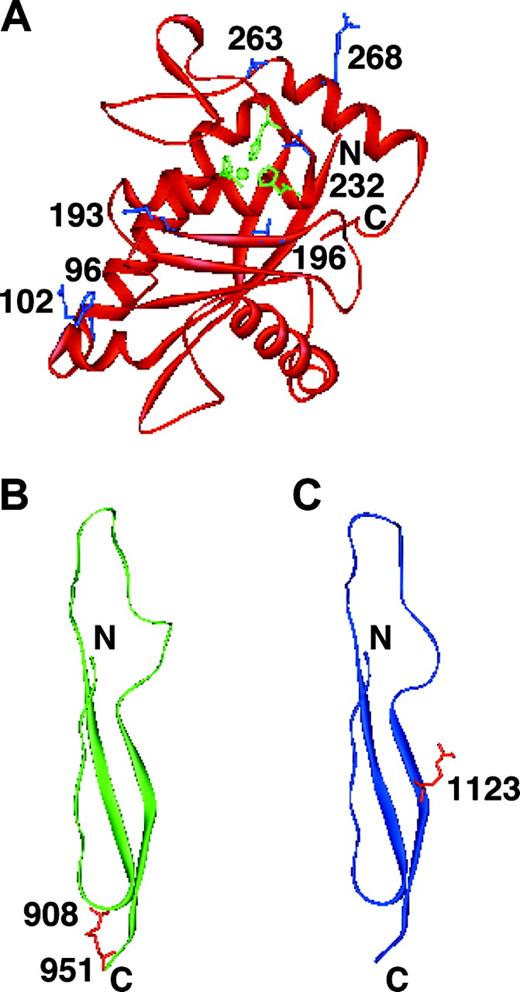
We constructed 3-dimensional molecular models of the metalloprotease and Tsp1 domains in which the mutations were identified. There are 2 missense mutations located in the metalloprotease domain: R193W identified in the present study and R268P identified in our previous study.18 The R193W mutation is located close to the active site. Even if the mutant protein is secreted, the mutation is predicted to disturb the activity, as shown in Figures 2, 3. The R268P mutation is located in the middle of an α-helix. It is well known that proline is an α-helix breaker. Therefore, it can be predicted that the R268P substitution would cause a secretion defect due to the disruption of one of the α-helices in the molecule.
The C908Y and R1123C mutations are present in the Tsp1-5 and Tsp1-8 domains, respectively (Figure 5B and 5C, respectively). The C908Y mutation may disrupt a potential disulfide bond and the R1123C mutation may create the mixed disulfide bond; both of them could disrupt the proper conformation of the enzyme, leading to the secretion defect.
Shown are 3-dimensional models of metalloprotease, Tsp1-5, and Tsp1-8 domains of ADAMTS13. (A) Metalloprotease domain. The side chains of Arg193 and Arg268 are shown in blue with residue numbers. The active site zinc ion coordinated by His224, His228, and His234 is also shown (green). The mutations of H96D, R102C, T196I, L232Q, and S263C reported by Levy et al12 and Schneppenheim et al31 are present in the metalloprotease domain. The side chains of His96, Arg102, Thr196, Leu232, and Ser263 are also shown in blue. (B) Tsp1-5 domain. The side chains of Cys908 and Cys951 are shown in red with residue numbers. (C) Tsp1-8 domain. The side chain of Arg1123 is shown in red with residue number.
Shown are 3-dimensional models of metalloprotease, Tsp1-5, and Tsp1-8 domains of ADAMTS13. (A) Metalloprotease domain. The side chains of Arg193 and Arg268 are shown in blue with residue numbers. The active site zinc ion coordinated by His224, His228, and His234 is also shown (green). The mutations of H96D, R102C, T196I, L232Q, and S263C reported by Levy et al12 and Schneppenheim et al31 are present in the metalloprotease domain. The side chains of His96, Arg102, Thr196, Leu232, and Ser263 are also shown in blue. (B) Tsp1-5 domain. The side chains of Cys908 and Cys951 are shown in red with residue numbers. (C) Tsp1-8 domain. The side chain of Arg1123 is shown in red with residue number.
Discussion
Here, we have identified 7 new mutations in the ADAMTS13 gene responsible for USS, including 4 missense mutations (R193W, I673F, C908Y, and R1123W) and 3 splice site mutations. We have reported 2 missense mutations (R268P and C508Y) and one nonsense mutation (Q449X).18 All 6 missense mutant proteins, except for the R193W mutant, were not secreted from the cells in the expression analysis using HeLa cells. Although the R193W mutant was secreted to some extent, it had little activity. This mutation may not cause a dramatic conformational change of the enzyme but may disrupt the active site locally.
To date, the analysis of the ADAMTS13 gene was reported in a total of 26 patients with congenital TTP, equivalent to USS, including those in the present study.12,18,31-33 All of the patients with congenital TTP have been shown to have ADAMTS13 gene mutations. Of these patients, 7 were homozygotes and 19 were compound heterozygotes. It is noteworthy that these mutation sites are distributed over exons 3 to 29 regardless of race. Among our 7 patients with USS, 2 are homozygotes, of whom 1 (patient C) has a family history of consanguineous marriage, and the other (patient B) appears not to have such a history. However, a careful routing analysis performed in this study revealed that 2 great-grandparents of patient B were from the same village in the Northeastern region of the Japanese mainland at the end of the 19th century. More interestingly, one Japanese patient with USS reported by Sasahara et al34 has the same homozygous mutation (Q449X) as our patient B (personal written communication from Drs David Ginsburg and Shigeru Tsuchiya, May 2003). This patient is a natural habitant of the same Northeastern region of Japan as patient B's ancestors. Our experience suggests that among USS patients consanguinity gives rise to the minor group with homozygous mutation of the ADAMTS13 gene, whereas the major group consists of individuals with compound heterozygous mutations not arising from consanguinity.
The most striking and earliest clinical sign of USS is a Coombs-test–negative severe hyperbilirubinemia soon after birth. In fact, all of our 7 patients had this history, and 6 of 7 were rescued by exchange blood transfusion and 1 by phototherapy. As noted by others35 and by us,16 some patients with USS, despite a severe lack of ADAMTS13 activity, have no acute episode of TTP/HUS during the newborn period, but develop the episode during early or late childhood. In some very unusual female cases, it has been said that the clinical manifestation is first noted after pregnancy.35 This is perhaps triggered by an increased amount of VWF together with a higher ratio of ultralarge or large VWF multimers than in the nonpregnant state. However, it has not been clarified whether this phenomenon is caused by overproduction of VWF or by a physiologic decrease of ADAMTS13 activity during the second and third trimesters of normal pregnancy, and/or by both. At present we cannot address why phenotypic differences on the clinical onset or signs exist. Besides pregnancy, it has been speculated that some other precipitating factors may include infections, cytokines, or chemical compounds.
In the present study, we constructed models of the metalloprotease domain, and Tsp1-5 and Tsp1-8 domains of ADAMTS13 using the coordinates of adamalysin II and the thrombospondin-1 type 1 repeats, respectively (Figure 5). The sequence alignments are highly important for obtaining the proper models. In the present study, the sequence alignments produced using PSI-BLAST were manually adjusted taking biologically important regions and secondary structure into consideration. Thus, the sequence identities of 22.1%, 28.8%, and 30.8% were obtained between the metalloprotease domain and adamalysin II, the Tsp1-5 domain and thrombospondin-1 type 1 repeats, and the Tsp1-8 domain and thrombospondin-1 type 1 repeats, respectively. Based on our experiences, the models constructed with the amino acid sequence identities higher than 20% are quite convincing. However, we did not construct the models of the cysteine-rich and spacer domains because their reference proteins were not obtained due to their unique sequences. In addition, the domain-domain interactions presumably present in the ADAMTS13 molecule are not predicted. Thus, the structure determined by the X-ray crystallography remains to be solved.
Prepublished online as Blood First Edition Paper, October 16, 2003; DOI 10.1182/blood-2003-06-1796.
Supported in part by Grants-in-Aid for Scientific Research grant from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (M. Matsumoto, K.K., T.M. and Y. Fujimura); from the Ministry of Health and Welfare of Japan for Blood Coagulation Abnormalities H14-02 (T.M. and Y. Fujimura); and from the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan (T.M. and K.K.).
M. Matsumoto and K.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs K. Kamide, S. Takiuchi, and Y. Kawano, Division of Hypertension and Nephrology, National Cardiovascular Center, for providing genomic DNAs isolated from individuals without TTP.