Abstract
The primary macrophage growth factor, colony-stimulating factor 1 (CSF-1), is expressed as a secreted glycoprotein or proteoglycan found in the circulation or as a biologically active cell surface glycoprotein (csCSF-1). To investigate the in vivo roles of csCSF-1, we created mice that exclusively express csCSF-1, in a normal tissue-specific and developmental manner, by transgenic expression of csCSF-1 in the CSF-1-deficient osteopetrotic (Csf1op/Csf1op) background. The gross defects of Csf1op/Csf1op mice, including growth retardation, failure of tooth eruption, and abnormal male and female reproductive functions were corrected. Macrophage densities in perinatal liver, bladder, sublinguinal salivary gland, kidney cortex, dermis, and synovial membrane were completely restored, whereas only partial or no restoration was achieved in adult liver, adrenal gland, kidney medulla, spleen, peritoneal cavity, and intestine. Residual osteopetrosis, significantly delayed trabecular bone resorption in the subepiphyseal region of the long bone, and incomplete correction of the hematologic abnormalities in the peripheral blood, bone marrow, and spleens of CSF-1-deficient mice were also found in mice exclusively expressing csCSF-1. These data suggest that although csCSF-1 alone is able to normalize several aspects of development in Csf1op/Csf1op mice, it cannot fully restore in vivo CSF-1 function, which requires the presence of the secreted glycoprotein and/or proteoglycan forms. (Blood. 2004;103:1114-1123)
Introduction
Colony-stimulating factor 1 (CSF-1), also known as macrophage CSF, is the primary regulator of the mononuclear phagocyte lineage and regulates cells of the female reproductive tract.1-6 All effects of CSF-1 are mediated by a high-affinity receptor tyrosine kinase7-10 encoded by the c-fms proto-oncogene.11 At least 5 mature human or mouse CSF-1 mRNAs (4.0 kb, 3.0 kb, 2.3 kb, 1.9 kb, and 1.6 kb) resulting from alternative splicing in exon 6 and the alternative usages of the 3′-untranslated region exons 9 and 10,12-18 have been shown to encode 3 isoforms of the CSF-1 protein: a secreted glycoprotein,19-21 a secreted proteoglycan,22,23 and a biologically active membrane-spanning cell surface glycoprotein18,24-29 (for a review, see Stanley30 ).
The primary source of the circulating proteoglycan and glycoprotein CSF-1 is thought to be the endothelial cells that line the small blood vessels (for a review, see Roth and Stanley31 ). CSF-1 is also synthesized locally,32 for example, by osteoblasts33,34 and by uterine epithelial cells.3 It has been suggested that regulation at particular tissue sites is mediated by local synthesis of the membrane-spanning, cell surface CSF-1 (csCSF-1), and/or selective sequestration of the secreted proteoglycan CSF-1 (spCSF-1).22,23,35
The csCSF-1 is encoded by a truncated mRNA in which part of the exon 6 sequence encoding the fragment containing the unique glycosaminoglycan addition site and the proteolytic cleavage sites used to release the secreted isoforms has been spliced out (Figure 1A).18,27 csCSF-1 is expressed in all cell types examined that express soluble CSF-1, including fibroblasts and osteoblasts.18,24-29,36,37 It has been shown to support macrophage proliferation25,38 and the formation of multinucleated osteoclastlike cells in vitro37,39,40 and in vivo.41 csCSF-1 was proposed to play adhesion molecule-like roles in its interaction with the CSF-1 receptor (CSF-1R) on leukemic cells in a culture system.42 When expressed on tumor cells, csCSF-1, but not secreted CSF-1, can induce tumor cell cytotoxicity by macrophages and also activate systemic immunity to the tumor cell.43-45 Glucocorticoids have been reported to enhance osteoclastogenesis by increasing the csCSF-1 level in osteoblasts.46 However, increased csCSF-1 expression did not enhance receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL)-driven osteoclastogenesis from marrow cells in another case.47
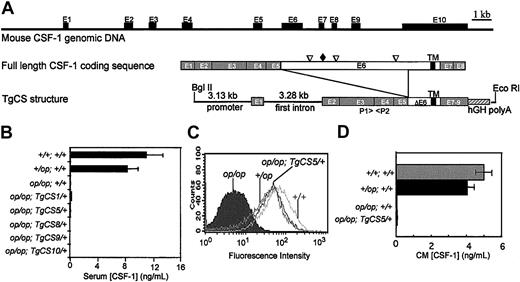
Mouse CSF-1 genomic structure, the TgCS transgene construct, and CSF-1 expression in Csf1op/Csf1op; TgCS/+ mice. (A) The genomic organization of the mouse CSF-1 gene constructed from the sequences of cDNA encoding full-length mouse CSF-1 (M21952), csCSF-1 (BC025593), and the genomic sequence of mouse chromosome 3 published in National Center for Biotechnology Information GenBank (NT_039239). The positions of 3 major proteolytic cleavage sites (▿), the unique glycosaminoglycan addition site (♦), and the transmembrane domain (TM) are shown in the coding sequence of full-length CSF-1. To construct the TgCS transgene, the exon 2-8 fragment of the cDNA encoding the csCSF-1 possessing a truncated exon 6 was cloned downstream of the 3.13-kb promoter and first intron fragment and an additional hGH polyA signal fragment was added at the 3′ end of the cDNA. The fragment of exon 6 deleted in the TgCS transgene encodes the peptide fragment containing the major proteolytic cleavage sites and the glycosaminoglycan addition site. BlgII and EcoRI were used to linearize TgCS plasmid DNA for microinjection. P1 and P2 were primers for PCR genotyping.32 (B) Serum CSF-1 concentrations of Csf1op/Csf1op; TgCS/+ mice and their controls measured by RIA. Means ± SD (n ≥ 5 mice). (C) Surface CSF-1 expression on skin fibroblasts derived from the mice of the indicated genotypes was measured by flow cytometry following staining with anti-CSF-1 antibodies. (D) The CSF-1 concentrations in culture supernatants of skin fibroblasts with the indicated genotypes were determined by RIA. Means ± SD of 3 cultures.
Mouse CSF-1 genomic structure, the TgCS transgene construct, and CSF-1 expression in Csf1op/Csf1op; TgCS/+ mice. (A) The genomic organization of the mouse CSF-1 gene constructed from the sequences of cDNA encoding full-length mouse CSF-1 (M21952), csCSF-1 (BC025593), and the genomic sequence of mouse chromosome 3 published in National Center for Biotechnology Information GenBank (NT_039239). The positions of 3 major proteolytic cleavage sites (▿), the unique glycosaminoglycan addition site (♦), and the transmembrane domain (TM) are shown in the coding sequence of full-length CSF-1. To construct the TgCS transgene, the exon 2-8 fragment of the cDNA encoding the csCSF-1 possessing a truncated exon 6 was cloned downstream of the 3.13-kb promoter and first intron fragment and an additional hGH polyA signal fragment was added at the 3′ end of the cDNA. The fragment of exon 6 deleted in the TgCS transgene encodes the peptide fragment containing the major proteolytic cleavage sites and the glycosaminoglycan addition site. BlgII and EcoRI were used to linearize TgCS plasmid DNA for microinjection. P1 and P2 were primers for PCR genotyping.32 (B) Serum CSF-1 concentrations of Csf1op/Csf1op; TgCS/+ mice and their controls measured by RIA. Means ± SD (n ≥ 5 mice). (C) Surface CSF-1 expression on skin fibroblasts derived from the mice of the indicated genotypes was measured by flow cytometry following staining with anti-CSF-1 antibodies. (D) The CSF-1 concentrations in culture supernatants of skin fibroblasts with the indicated genotypes were determined by RIA. Means ± SD of 3 cultures.
Osteopetrotic (Csf1op/Csf1op) mice harbor an inactivating mutation in the coding region of the CSF-1 gene and are CSF-1 deficient.48-50 They are osteopetrotic due to their paucity of osteoclasts.51 They are toothless, have low body weight and low growth rate, and are deficient in tissue macrophages.49,51-55 In addition, they have defects in both male and female fertility and in neural development (for a review, see Pollard and Stanley5 ). Because CSF-1R expression outside the female reproductive tract is largely restricted to mononuclear phagocytes (for reviews, see Pollard and Stanley5 and Stanley30 ), the pleiotropic phenotype of the Csf1op/Csf1op mouse is apparently due to a reduction in trophic and/or scavenger functions of the tissue macrophages regulated by CSF-1, secondary to the reduction of their concentration in tissues.55
Reconstitution of circulating levels of CSF-1 in Csf1op/Csf1op mice, achieved by administration of recombinant human CSF-1 (rhCSF-1), resulted in correction of several defects in Csf1op/Csf1op mice.32,50,55-57 Expression in Csf1op/Csf1op mice of a transgene encoding the full length CSF-1 precursor driven by a 3.13-kb CSF-1 gene promoter and the first intron reconstituted both circulating and tissue levels of secreted CSF-1 and some csCSF-1 expression in a normal tissue-specific and developmental pattern. This transgene was able to completely correct the reported defects in Csf1op/Csf1op mice.32 Here we have used the same CSF-1 promoter and first intron fragment to drive the expression of the cDNA encoding the csCSF-1 on the Csf1op/Csf1op background. Multiple csCSF-1 transgenic lines, expressing wild-type levels of csCSF-1 on their fibroblasts and with no detectable circulating CSF-1, were established. Transgenic expression of csCSF-1 completely restored some defects of Csf1op/Csf1op mice, including growth retardation, defective tooth eruption and abnormal reproductive functions, as well as the deficiencies of F4/80+ tissue macrophages in bladder, sublinguinal salivary gland, kidney cortex, dermis, and synovial membrane. However, macrophage densities in liver, adrenal, spleen, and large intestine were only partially restored. Residual osteopetrosis, delayed trabecular bone resorption in the subepiphyseal region of the long bone, and abnormal hematologic parameters were also found in these mice.
Materials and methods
Mice
Osteopetrotic Csf1op/Csf1op mice, CSF-1 transgenic Csf1op/Csf1op mice, and wild-type controls (Csf1+/Csf1+or Csf1+/Csf1op) were bred and maintained on an outbred background (C57BL/J × C3Heb/FeJ-a/a × CD1) or on an FVB/NJ background onto which the Csf1op mutation had been backcrossed for at least 10 generations (Table 1), behind a barrier in the Institute for Animal Studies of the Albert Einstein College of Medicine. Csf1op/Csf1op mice were identified by the absence of incisor eruption at 21 days of age and were fed a powdered mixture of mouse chow ad libitum and infant formula (Enfamil) daily. Transgenic mice and control mice received regular mouse chow. Reciprocal mating schemes (female Csf1op/Csf1op;TgCS/+ × male Csf1+/Csf1op; +/+ or female Csf1+/Csf1op; +/+ × male Csf1op/Csf1op; TgCS/+) were used to maintain transgenic lines and generate experimental animals.
Transgene constructs, production of transgenic animals, and genotyping
To assemble the csCSF-1 transgene construct, a 734-bp exon 2-8 cDNA fragment encoding the csCSF-1 isoform was used to replace the corresponding portion in the full-length CSF-1 transgene construct.32 This resulted in the deletion of the sequence in exon 6 encoding amino acids 182-476 of the full-length sequence that encodes the proteolytic cleavage sites and the glycosaminoglycan addition site (Figure 1A). The preparation of linearized DNA for microinjection, production of transgenic mice, and the genotyping of mice bearing the Csf1op allele or the transgene or both were carried out as described for the full-length CSF-1 transgenic (TgC) mice.32 According to the “Mouse Nomenclature Rules and Guidelines” published by the MGI Nomenclature (http://www.informatics.jax.org/mgihome/nomen/), the csCSF-1 transgenes were designated as TgN(CSCsf1)Ers (abbreviated to TgCS).
Measurement of CSF-1
The CSF-1 concentrations in sera, tissues, and in the media conditioned by skin fibroblasts were measured by mouse CSF-1 radioimmunoassay (RIA).2,58 Tissue extracts were prepared as described previously.32 Detection of CSF-1 in the tissue extracts of Csf1op/Csf1op; TgCS/+ mice is probably due to the release of csCSF-1 to the soluble fraction, because the extraction procedure involves homogenization and heating (56°C, 30 minutes) and csCSF-1 is sensitive to cleavage by trypsin and is released in a biologically active form, detected by the RIA.32 csCSF-1 on cultured fibroblasts was measured by flow cytometry (see “Immunohistochemistry, histochemistry, and flow cytometry”).
X-radiographic analysis of mouse skeletal structure
Radiographs were produced by exposing killed or anesthetized mice in a Faxitron pathology specimen x-ray cabinet (Faxitron X-Ray, Buffalo Grove, IL). The animals were posed immediately above a fine-grained Polaroid 665 instant negative film package. Exposure was set at 90 kV for 2 minutes. The negatives were developed and printed according to the manufacturer's instructions (Polaroid, Cambridge, MA).
Immunohistochemistry, histochemistry, and flow cytometry
For immunostaining with rat monoclonal antibody F4/8059 and histochemical localization of tartrate-resistant acid phosphatase (TRAP), siblings of the different genotypes were perfused and tissues fixed, decalcified (knee joint only), embedded, sectioned, and immunostained as described.55 F4/80+ cells in tissue sections of at least 2 mice of a particular genotype at each age were quantitated as described.55 The scorer of F4/80+ cell numbers was blinded to the genotypes of sections. Frozen sections of spleen taken from 1-month-old mice were stained for marginal metallophilic macrophages using the rat monoclonal antibody, MOMA-1.60 Whole-mount preparations of the fourth inguinal mammary gland were stained with alum carmine as described.61 For flow cytometry analysis of csCSF-1, primary skin fibroblasts were detached from Csf1+/Csf1+; +/+, Csf1+/Csf1op; +/+, Csf1op/Csf1op; +/+, and Csf1op/Csf1op; TgCS5/+ cultures by treatment of the monolayers with 2 mM EDTA (ethylenediaminetetraacetic acid) in phosphate-buffered saline and the single-cell suspensions incubated sequentially with biotinylated F(ab′)2 fragments of rat antimouse CSF-1 YYG106 monoclonal antibody2 followed by phycoerythrin (PE)-streptavidin (PharMingen, San Diego, CA). For cell surface marker studies, red cell lysates of heparinized blood, bone marrow cells, and splenic cell samples, antibody staining (fluorescein isothiocyanate [FITC]-CD45.2, peridinin chlorophyll protein [PerCp]-B220, PE-Gr1, FITC-CD11b, PE-Ter119, PE-CD3; PharMingen), fluorescence-activated cell sorting (FACS), and data analysis of FACS analyses were carried out using standard procedures with a Becton Dickinson FACSCalibur (San Jose, CA) in the FACS Facility of Albert Einstein College of Medicine.
Statistical analysis
The means and SDs of all numeric data were calculated. Data were analyzed statistically using the Student t test. Differences were considered statistically significant for comparisons of data sets yielding P ≤ .05.
Results
Csf1op/Csf1op; TgCS/+ mice express normal levels of csCSF-1 and no detectable circulating CSF-1
Our previous results indicate that the fragment of the mouse CSF-1 gene containing 3.13 kb of the promoter and entire first intron confers an essentially wild-type CSF-1 expression pattern in mice.32 The same fragment was used to drive the expression of a cDNA encoding the csCSF-1 with the same human growth hormone polyriboadenylic acid (hGH polyA) addition signal sequence at the 3′ end in the creation of TgCS transgene (Figure 1). A summary of the basic phenotypic characteristics of the 10 TgN(CSCSF1)Ers1-10 (TgCS1-10) mouse lines obtained is presented in Table 1. Whereas all 10 founder mice bred normally and produced normal numbers of progeny, only 6 were found to transmit the transgene. Csf1op/Csf1op; TgCS/+ mice were obtained for 5 of these lines (TgCS1, TgCS5, TgCS8, TgCS9, TgCS10; Table 1). Results initially collected from TgCS1 and TgCS5 lines on the outbred background were indistinguishable from subsequent data obtained from Csf1op/Csf1op; TgCS5/+ mice on the FVB/NJ inbred background on to which both the Csf1op and TgCS5 alleles had been backcrossed for more than 5 generations. TgCS8, TgCS9, and TgCS10 lines were generated and maintained on the FVB/NJ background. The phenotypes of mice from all Csf1op/Csf1op; TgCS/+ lines were similar, excluding the possibility that they resulted from random transgene insertion-related positional effects or insertional mutagenesis by the transgene.
No detectable CSF-1 was found in the sera of mice of all 5 Csf1op/Csf1op; TgCS/+ lines using an RIA that detects only biologically active CSF-12 (Figure 1B). Extractable tissue CSF-1 levels in most of the tissues from Csf1op/Csf1op; TgCS5/+ mice were similar to their controls from wild-type mice, probably due to the efficient solubilization of csCSF-1 by cleavage during extraction (data not shown). To measure the relative levels of csCSF-1 expression, primary skin fibroblasts isolated from Csf1+/Csf1+, Csf1+/Csf1op, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice were cultured in vitro, stained with a rat antimouse CSF-1 antibody, and analyzed by flow cytometry (Figure 1C). Levels of csCSF-1 expression on Csf1op/Csf1op; TgCS5/+ fibroblasts were similar to the levels on Csf1+/Csf1op fibroblasts and less than the levels on Csf1+/Csf1+ fibroblasts. In addition, no CSF-1 was detected in the medium conditioned by Csf1op/Csf1op; TgCS5/+ fibroblasts (Figure 1D), further confirming their exclusive expression of csCSF-1.
Normalized growth rate, male and female reproductive function, and incisor eruption in Csf1op/Csf1op; TgCS/+ mice
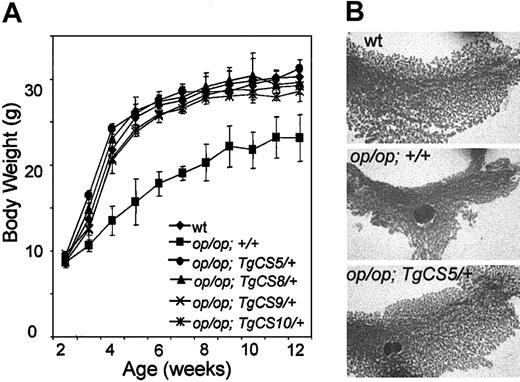
Apart from their severe osteopetrosis, Csf1op/Csf1op mice are toothless, have impaired mammary gland development, and have both female and male reproductive defects.5,6,51 Despite their powdered chow and milk formula diet, these mice still exhibit a decreased adult body weight and a markedly retarded growth rate (Figure 2A). We previously showed that the full-length transgene, TgC, is able to fully restore the growth retardation in Csf1op/Csf1op; TgC/+ mice.32 Similarly, there was no difference in the body weight of mice exclusively expressing csCSF-1 and their wild-type littermate controls, and the growth rate of Csf1op/Csf1op; TgCS/+ mice derived from all the TgCS lines was normalized (Figure 2A) as was incisor eruption (Table 1).
Normal growth rate, adult body weight, and mammary gland in development in Csf1op/Csf1op; TgCS5/+ mice. (A) Groups of male mice (n ≥ 5) of each of the indicated genotypes were weighed at weekly intervals from 2 to 12 weeks of age. Means ± SD. (B) Whole-mount alum-carmine staining of the fourth inguinal mammary gland from pregnant mice of the indicated genotypes at 18 days of gestation (original magnification × 2.6).
Normal growth rate, adult body weight, and mammary gland in development in Csf1op/Csf1op; TgCS5/+ mice. (A) Groups of male mice (n ≥ 5) of each of the indicated genotypes were weighed at weekly intervals from 2 to 12 weeks of age. Means ± SD. (B) Whole-mount alum-carmine staining of the fourth inguinal mammary gland from pregnant mice of the indicated genotypes at 18 days of gestation (original magnification × 2.6).
The important role of CSF-1 in ovulation, preimplantation, placental function, regulation of the estrous cycle, and lactation has been described previously (for reviews, see Pollard and Stanley5 and Cohen et al6 ). Both the CSF-1-deficient Csf1op/Csf1op mice and the CSF-1R-null mice have increased estrous cycle times, a failure of mammary gland development during pregnancy, and a decreased male libido.10,62 In contrast to Csf1op/Csf1op mice, female Csf1op/Csf1op; TgCS/+ mice were able to nurture their progeny normally and whole-mount alum carmine staining of mammary glands from 18-day pregnant Csf1op/Csf1op; TgCS5/+ mice showed normal mammary gland development (Figure 2B).
To further test the reproductive function of Csf1op/Csf1op; TgCS5/+ mice, we carried out reciprocal matings of Csf1op/Csf1op; TgCS5/+ mice with Csf1+/Csf1op;+/+ mice (Table 2). The results of these crosses indicate that (1) the litter sizes from crosses involving either Csf1op/Csf1op; TgCS5/+ males or females and Csf1op/+ mice were similar, (2) the genotypic percentages were as expected from mendelian segregation of the transgene and the lower survival of Csf1op/Csf1op mice at the time of genotyping (3 weeks), (3) the percentage of Csf1op/Csf1op;+/+ mice produced by mating pairs with female Csf1op/Csf1op; TgCS5/+ and Csf1op/Csf1op; TgCS10/+ mice were lower than the reciprocal mating pairs with wild-type (Csf1op/+) females (5% versus 17.3% and 5% versus 12.6%, respectively). The reasons for this effect are not clear.
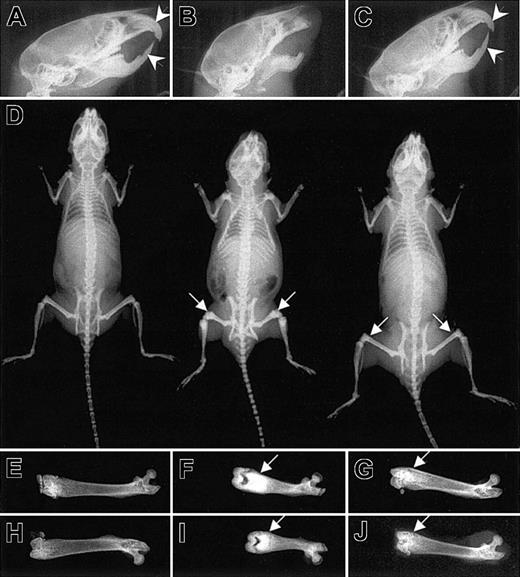
Incisor eruption in Csf1op/Csf1op mice was restored either by daily subcutaneous injection of rhCSF-155 or the normal tissue-specific and developmental expression of full-length CSF-1.32 In Csf1op/Csf1op; TgCS/+ mice, upper and lower incisor eruptions were indistinguishable from those of their wild-type littermates, occurring on postnatal day 9 to 10. Although incisor teeth appeared abnormally curved or misaligned in the rhCSF-1-injected Csf1op/Csf1op mice,32 as in the case of Csf1op/Csf1op; TgC/+ mice,32 the incisor teeth were always normal and properly aligned in Csf1op/Csf1op; TgCS/+ mice (Figure 3A-C).
Normal incisor tooth eruption but residual osteopetrosis in the Csf1op/Csf1op; TgCS5/+ mice. Radiograms of 8-week-old wild-type, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice. (A-C) Normal incisor eruptions (arrowheads) in Csf1op/Csf1op; TgCS5/+ mice. (D) Radiographs of whole bodies of wild-type (left), Csf1op/Csf1op (middle), and Csf1op/Csf1op; TgCS5/+ mice (right). (E-J) Radiographs of individual femurs from 2 different wild-type (E,H), Csf1op/Csf1op (F,I) and Csf1op/Csf1op; TgCS5/+ mice (G, J). Arrows in panels D, F, G, I, and J point to the areas of increased bone density.
Normal incisor tooth eruption but residual osteopetrosis in the Csf1op/Csf1op; TgCS5/+ mice. Radiograms of 8-week-old wild-type, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice. (A-C) Normal incisor eruptions (arrowheads) in Csf1op/Csf1op; TgCS5/+ mice. (D) Radiographs of whole bodies of wild-type (left), Csf1op/Csf1op (middle), and Csf1op/Csf1op; TgCS5/+ mice (right). (E-J) Radiographs of individual femurs from 2 different wild-type (E,H), Csf1op/Csf1op (F,I) and Csf1op/Csf1op; TgCS5/+ mice (G, J). Arrows in panels D, F, G, I, and J point to the areas of increased bone density.
Residual osteopetrosis and delayed trabecular bone resorption in Csf1op/Csf1op; TgCS/+ mice
CSF-1-deficient Csf1op/Csf1op mice exhibit impaired bone resorption associated with a paucity of osteoclasts.51 Their inability to remodel bone results in skeletal deformities. For example, the long bones are dense, short, and thick (Figure 3D, middle image, and F,I) and their flat bony plates produce a characteristically domed skull (Figure 3B). The decreased head size, shortening of the facial bones, smaller overall skeletal size, and abnormal shape of femurs in the Csf1op/Csf1op mice were corrected by TgCS expression (Figure 3C-D). However, there was residual increased bone density in the femurs and tibias in the 2-month-old Csf1op/Csf1op; TgCS5/+ mice (Figure 3D,E-J). X-radiography of 1-week-old, 2-week-old, 1-month-old, and 3-month-old Csf1op/Csf1op; TgCS/+ mice derived from all TgCS lines revealed a similar residual osteopetrosis in the long bones (data not shown).
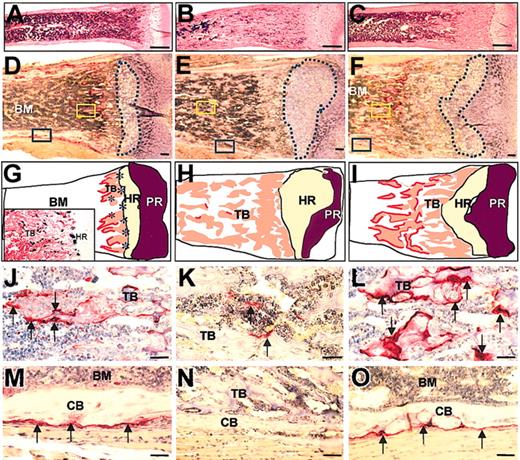
To investigate the bone resorption, marrow cavity formation, and osteoclastogenesis in Csf1op/Csf1op; TgCS/+ mice, femurs from 2-day, 1-week, 2-week, and 1-month old Csf1op/Csf1op; TgCS/+ mice derived from all TgCS lines and their wild-type and Csf1op/Csf1op;+/+ littermate controls were sectioned and subjected to TRAP staining and hematoxylin counterstaining. Formation of the marrow cavity initiated normally in the femurs of 2 day-old Csf1op/Csf1op; TgCS/+ mice. However, the resorption of trabecular bone in the subepiphyseal region was significantly delayed in Csf1op/Csf1op; TgCS/+ mice, witnessed by the increased distance from the trabecular bone resorption edge to the edge of the chondrocyte hypertrophic region, compared with wild-type femurs in 2-week-old mice (Figure 4A-F). In addition, the expanded cartilage region, including both proliferative and hypertrophic chondrocyte regions, characteristic of Csf1op/Csf1op mice in both radiographs (Figure 3F,I) and histologic sections, were also seen in Csf1op/Csf1op; TgCS/+ mice and, although less severe, were especially apparent in the earlier stages of bone development (data not shown). TRAP+ osteoclasts were dramatically increased in number and size in Csf1op/Csf1op; TgCS/+ femurs (Figure 4L,O) compared with Csf1op/Csf1op femurs (Figure 4K,N). Similar to wild-type femurs, TRAP+ osteoclasts in Csf1op/Csf1op; TgCS/+ femurs were large and fully expanded and covered virtually all the free trabecular bone and periosteal surfaces although the free trabecular surface in the wild-type femurs was significantly less than the free trabecular surface of Csf1op/Csf1op; TgCS/+ femurs. This may explain the smaller or less expanded appearance of TRAP+ cells in the wild-type femurs (Figure 4J,M). These data indicate that local csCSF-1 expression by osteoblasts and/or bone marrow stromal cells32 is sufficient to support the osteoclastogenesis, but that the rate of trabecular bone resorption by osteoclasts in Csf1op/Csf1op; TgCS/+ femurs is slower.
Increased trabecular bone in the subepiphyseal region and normal TRAP+ osteoclast numbers in femurs of 2-week-old Csf1op/Csf1op; TgCS5/+ mice. (A-F) Images of sections of the subepiphyseal regions of the distal femurs from wild-type (A,D), Csf1op/Csf1op (B,E), and Csf1op/Csf1op; TgCS5/+ mice (C,F), stained for TRAP and counterstained with hematoxylin, show the larger amount of unresorbed trabecular bone and smaller bone marrow cavity of the femurs of Csf1op/Csf1op; TgCS5/+ mice compared with those of wild-type mice. (D-F) Regions circled by the dashed blue lines delineate hypertrophic chondrocyte regions. (G-I) Illustration of images in panels D-F shows bone marrow (BM) cavity, trabecular bone (TB), hypertrophic region (HR) of cartilage, proliferative region (PR) of cartilage, and osteoclasts as the red outline of the trabecular bone. Asterisks in HR indicate concentration of CSF-1-expressing cells visualized in CSF-1-promoter-first intron driven lac Z (TgZ) transgenic mice32 and shown in the insert to G. (J-O) High magnification images of regions of yellow (J-L) and blue (M-O) boxed areas in panels D-F, showing the TRAP+ osteoclasts in the regions of trabecular bone (J-L) and periosteum (M-O) in wild-type (J,M), Csf1op/Csf1op (K,N), and Csf1op/Csf1op; TgCS5/+ mice (L,O). TRAP+ osteoclasts (arrows) are stained red. CB indicates cortical bone; bar is 1 mm in panels A-C and 50 μm in panels D-O.
Increased trabecular bone in the subepiphyseal region and normal TRAP+ osteoclast numbers in femurs of 2-week-old Csf1op/Csf1op; TgCS5/+ mice. (A-F) Images of sections of the subepiphyseal regions of the distal femurs from wild-type (A,D), Csf1op/Csf1op (B,E), and Csf1op/Csf1op; TgCS5/+ mice (C,F), stained for TRAP and counterstained with hematoxylin, show the larger amount of unresorbed trabecular bone and smaller bone marrow cavity of the femurs of Csf1op/Csf1op; TgCS5/+ mice compared with those of wild-type mice. (D-F) Regions circled by the dashed blue lines delineate hypertrophic chondrocyte regions. (G-I) Illustration of images in panels D-F shows bone marrow (BM) cavity, trabecular bone (TB), hypertrophic region (HR) of cartilage, proliferative region (PR) of cartilage, and osteoclasts as the red outline of the trabecular bone. Asterisks in HR indicate concentration of CSF-1-expressing cells visualized in CSF-1-promoter-first intron driven lac Z (TgZ) transgenic mice32 and shown in the insert to G. (J-O) High magnification images of regions of yellow (J-L) and blue (M-O) boxed areas in panels D-F, showing the TRAP+ osteoclasts in the regions of trabecular bone (J-L) and periosteum (M-O) in wild-type (J,M), Csf1op/Csf1op (K,N), and Csf1op/Csf1op; TgCS5/+ mice (L,O). TRAP+ osteoclasts (arrows) are stained red. CB indicates cortical bone; bar is 1 mm in panels A-C and 50 μm in panels D-O.
Incomplete restoration of tissue macrophage densities in Csf1op/Csf1op; TgCS/+ mice
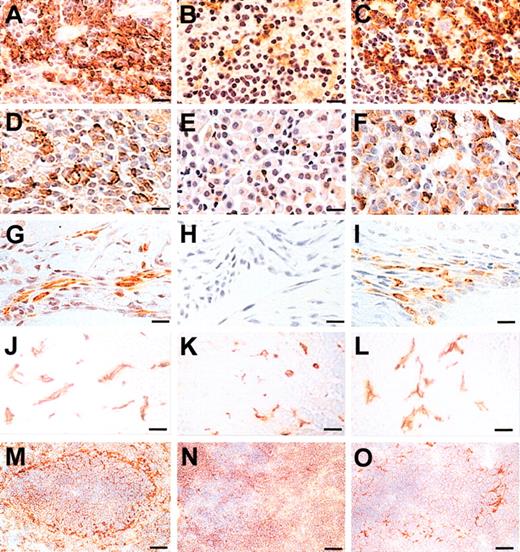
CSF-1 is the primary regulator of the differentiation, proliferation, and survival of mononuclear phagocytes. Defects in tissue macrophage development are among the primary defects found in CSF-1-deficient Csf1op/Csf1op mice.55 F4/80 is a well-recognized macrophage-specific marker and a rat antimouse F4/80 monoclonal antibody has been widely used to evaluate tissue macrophage populations.63 Most F4/80+ macrophage populations are either partially or completely dependent on CSF-1 for their development and maintenance.55 Complete restoration of tissue macrophages was achieved by normal tissue-specific and developmental expression of a full-length CSF-1 transgene, TgC,32 whereas daily postnatal injection of rhCSF-1 at concentrations sufficient to maintain circulating CSF-1 at above normal concentrations was unable to restore the F4/80+ macrophages that colonize the adrenal gland and dense connective tissues including tendon, striated muscle, deep dermis, synovium, and periosteum, but successfully restored the macrophages in other tissues.55 To examine the restoration of tissue macrophages by transgenic expression of TgCS in Csf1op/Csf1op mice, F4/80+ macrophages were stained in sections of various tissues dissected at the age at which highest macrophage densities were observed during the course of postnatal development.55 Macrophage morphology of Csf1op/Csf1op; TgCS5/+ tissues revealed by F4/80 staining was essentially comparable with the staining in wild-type tissues (Figure 5). F4/80+ macrophages in dermis, synovial membrane, bladder, salivary gland, kidney cortex, and 2-day liver were restored to the wild-type density by TgCS expression (Table 3; Figure 5G-I). However, the F4/80+ macrophage densities in spleen, adrenal gland, large intestine, and kidney medulla were only partially corrected (Table 3; Figure 5A-F). Interestingly, in contrast to 2-day liver, macrophage densities in 3-month old Csf1op/Csf1op liver were unaffected by TgCS expression (Table 3). However, TgCS expression markedly restored the wild-type, dendritic morphology of these macrophages (Figure 5J-L). The MOMA-1+ metallophilic macrophages surrounding the white pulp of spleen were also only partially restored in the Csf1op/Csf1op; TgCS5/+ mice (Figure 5 M-O).
MOMA-1+ and F4/80+ macrophages in tissues of Csf1op/Csf1op; TgCS5/+ mice. (A-L) Paraffin-embedded sections of spleens (A-C), adrenal medulla (D-F), synovial membrane (G-I), and liver (J-L) from 3-month-old wild-type (A,D,G,J), Csf1op/Csf1op (B,E,H,K), and Csf1op/Csf1op; TgCS/+ mice (C,F,I,L) stained with the macrophage-specific anti-F4/80 antibody and counterstained (A-I) with hematoxylin. (M-O) Frozen spleen sections from 1-month-old wild-type (M), Csf1op/Csf1op (N), and Csf1op/Csf1op; TgCS/+ mice (O) stained with the marginal metallophilic macrophage-specific monoclonal antibody, MOMA-1, and counterstained with hematoxylin. MOMA-1+ brown dendritic cells surround the white pulp in spleen. Bars: 50 μm (A-L) and 80 μm (M-O).
MOMA-1+ and F4/80+ macrophages in tissues of Csf1op/Csf1op; TgCS5/+ mice. (A-L) Paraffin-embedded sections of spleens (A-C), adrenal medulla (D-F), synovial membrane (G-I), and liver (J-L) from 3-month-old wild-type (A,D,G,J), Csf1op/Csf1op (B,E,H,K), and Csf1op/Csf1op; TgCS/+ mice (C,F,I,L) stained with the macrophage-specific anti-F4/80 antibody and counterstained (A-I) with hematoxylin. (M-O) Frozen spleen sections from 1-month-old wild-type (M), Csf1op/Csf1op (N), and Csf1op/Csf1op; TgCS/+ mice (O) stained with the marginal metallophilic macrophage-specific monoclonal antibody, MOMA-1, and counterstained with hematoxylin. MOMA-1+ brown dendritic cells surround the white pulp in spleen. Bars: 50 μm (A-L) and 80 μm (M-O).
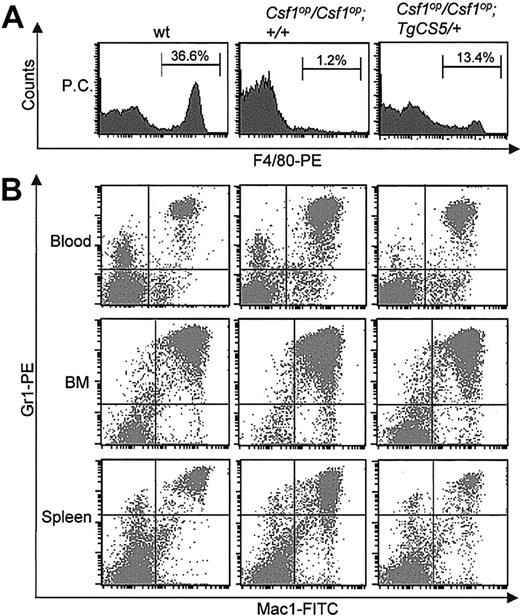
The peritoneal cavity cellularity of Csf1op/Csf1op mice (0.2 ± 0.11 × 106 cells/mouse) was also only partially restored by expression of TgCS (0.54 ± 0.13 × 106 cells/mouse), the Csf1op/Csf1op; TgCS/+ cellularity being significantly lower than the cellularity in wild-type mice (1.67 ± 0.33 × 106 cells/mouse). In particular, the percentage of F4/80+ macrophages in the peritoneal cavity was increased by TgCS expression but was still significantly lower than in wild-type control mice (Figure 6A).
Myeloid lineage parameters in Csf1op/Csf1op; TgCS5/+ mice. (A) Representative histograms of FACS analyses of F4/80-PE antibody-stained peritoneal cavity (PC) cells from wild-type, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice. Means ± SD (n = 3 mice) of the percentages of F4/80+ cells are indicated. (B) Representative dot plots of the FACS analyses of single-cell suspensions of peripheral blood, bone marrow (BM), and splenic cells from wild-type, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice double-stained with Gr1-PE and Mac1-FITC antibodies and analyzed by FACS. The means and percentages of such an analysis for 3 or more mice of each genotype are presented in Table 4.
Myeloid lineage parameters in Csf1op/Csf1op; TgCS5/+ mice. (A) Representative histograms of FACS analyses of F4/80-PE antibody-stained peritoneal cavity (PC) cells from wild-type, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice. Means ± SD (n = 3 mice) of the percentages of F4/80+ cells are indicated. (B) Representative dot plots of the FACS analyses of single-cell suspensions of peripheral blood, bone marrow (BM), and splenic cells from wild-type, Csf1op/Csf1op, and Csf1op/Csf1op; TgCS5/+ mice double-stained with Gr1-PE and Mac1-FITC antibodies and analyzed by FACS. The means and percentages of such an analysis for 3 or more mice of each genotype are presented in Table 4.
Abnormal hematologic parameters in Csf1op/Csf1op; TgCS/+ mice
Beside the well-known deficiency of tissue macrophages, the hematologic defects of Csf1op/Csf1op mice include a compensatory extramedullary splenic hematopoiesis in young mice, resulting from the reduced bone marrow cellularity, together with an increase in granulocytopoiesis and a decrease in B lymphopoiesis. To examine the effects of TgCS on the hematologic abnormalities of Csf1op/Csf1op mice, we analyzed total cellularity and lineage marker expression of peripheral blood, bone marrow, and spleens of 2-month-old Csf1op/Csf1op; TgCS5/+, wild-type and Csf1op/Csf1op mice. The bone marrow and peripheral blood white cell concentrations of Csf1op/Csf1op; TgCS/+ mice were lower than wild-type levels and were not significantly different from the concentrations in Csf1op/Csf1op mice (Table 4). In addition, splenic cellularity was significantly elevated compared with wild-type and not significantly different from Csf1op/Csf1op, indicating that compensatory extramedullary hematopoiesis also occurs in the Csf1op/Csf1op; TgCS/+ mice. The failure of TgCS expression to restore hematopoiesis is evidenced by the incomplete recovery of the B-cell deficiency in blood and bone marrow (B220+ cells; Table 4) and the increased granulocyte (Gr1+) numbers in peripheral blood (Gr1+; Figure 6B; Table 4). As expected, there was no significant difference in T-cell (CD3+) markers in the peripheral blood, bone marrow, and spleen between mice of the 3 genotypes. Interestingly, the populations of granulocytes in the bone marrow and spleen of Csf1op/Csf1op; TgCS5/+ mice were significantly lower than in wild-type mice whose levels were lower than those in Csf1op/Csf1op mice, suggesting a possible negative effect of csCSF-1 on granulocytopoiesis. Interestingly, our data did not show any significant difference of the Mac1+/Gr-1- population, commonly described as monocytes, in peripheral blood, bone marrow, and spleen among the wild-type, Csf1op/Csf1op and Csf1op/Csf1op; TgCS5/+ mice (Figure 6B; Table 4). Similar results were obtained when PE-F4/80 antibody staining was used to detect monocytes in blood (data not shown). The failure to detect a decrease in blood monocyte levels in Csf1op/Csf1op compared with wild-type mice is consistent with some previous reports55,64,65 but differs from others.32,52,66 The confirmation of the Mac1/Gr1 staining results by staining for the relatively specific mononuclear phagocyte marker, F4/80, indicates that blood monocyte levels in 2-month-old wild-type, Csf1op/Csf1op and Csf1op/Csf1op; TgCS/+ mice do not differ significantly.
Discussion
The secreted proteoglycan, secreted glycoprotein, and membrane-spanning cell surface isoforms of CSF-1 are produced from a single-copy CSF-1 gene by differential mRNA splicing and posttranslational modification. Although the general role of CSF-1 as the primary regulator of mononuclear phagocytic lineage development has been established both in vitro and in vivo, the specific biologic functions of each CSF-1 isoform have not been defined. csCSF-1 is stably expressed at the cell surface where it is slowly and inefficiently cleaved to yield a soluble form of the growth factor.22,27,67 In vitro experiments have shown that it is biologically active as an extracted membrane protein28 and that it is active in situ for macrophage proliferation,25,68-70 for myelopoiesis,38,69 and for osteoclastogenesis.29,39,40 Recently, a csCSF-1 transgene driven by the collagen Iα promoter has been shown to correct the bone density and tooth eruption defects of Csf1op/Csf1op mice.41
In previous studies, we identified a 3.13-kb promoter and first intron fragment of mouse CSF-1 gene that conferred an essentially normal tissue-specific and developmental CSF-1 expression pattern. This fragment was successfully used to drive the transgenic expression of the full-length CSF-1 gene (TgC), which encodes CSF-1 proteoglycan, glycoprotein, and some csCSF-1, in Csf1op/Csf1op mice. By comparing the effects of postnatal restoration of circulating CSF-1 by daily subcutaneous injection of rhCSF-1 with transgenic expression of full length CSF-1, we concluded that the local production of CSF-1 is required to completely restore tissue macrophage density in most Csf1op/Csf1op mouse tissues.32 In the present study, we have characterized the phenotypes of Csf1op/Csf1op; TgCS/+ mice in which csCSF-1 was exclusively expressed under the control of the CSF-1 promoter/first intron fragment. Similar to the reconstitution of gross defects in Csf1op/Csf1op mice by TgC,32 TgCS was also able to correct the several gross defects including the tooth eruption, growth retardation, and male and female reproductive function. However, in contrast to TgC, we found that TgCS was not able to fully restore the osteoclastic bone resorption, the deficiency of macrophages in some tissues, and the hematologic abnormalities in Csf1op/Csf1op mice. A comparison of the restoration of the Csf1op/Csf1op defects by daily injection of rhCSF-1 or expression of the TgC or TgCS transgenes is summarized in Table 5.
To ensure that the phenotype of Csf1op/Csf1op; TgCS/+ mice reflects the true expression pattern of csCSF-1 in vivo, we used a well-established transgene driver that confers normal tissue-specific expression in a copy number-dependent, position-independent manner,32 and we have characterized 5 independent transgenic lines. Similar results were obtained for each TgCS line for all of the phenotypic characteristics examined. All Csf1op/Csf1op; TgCS/+ mouse lines failed to express circulating CSF-1, and their fibroblasts failed to produce significant soluble CSF-1, but expressed wild-type levels of csCSF-1. Furthermore, a similar phenotype was observed for Csf1op/Csf1op; TgCS5/+ mice on both outbred and FVB/NJ backgrounds. These results eliminate the possibility of effects of other CSF-1 isoforms, of genetic background, or of transgene position in these Csf1op/Csf1op; TgCS/+ mice.
In contrast to Csf1op/Csf1op mice injected daily with rhCSF-1, in which circulating CSF-1 was restored and in which the Csf1op/Csf1op defects were only partially corrected, Csf1op/Csf1op; TgC/+ mice were completely corrected,32 indicating that the full-length transgene, which expresses all 3 CSF-1 isoforms, provided the required embryonic, humoral, and local regulation. Transgenic expression of TgCS significantly corrected many of the defects of Csf1op/Csf1op mice, including macrophage densities in several tissues. In addition, the morphology of F4/80+ or MOMA-1+ tissue macrophages (Figure 5) and TRAP+ osteoclasts (Figure 4) in Csf1op/Csf1op; TgCS5/+ mice have a dendritic or spread morphology like wild-type cells, whereas these cells were generally smaller, rounder, and less spread in the Csf1op/Csf1op mice. These observations indicate that in a qualitative sense, csCSF-1 was able to support the generation of the fully differentiated tissue macrophages and osteoclasts. They also suggest that csCSF-1, either via direct cell-cell interaction, or after its local release, determines the dendritic, spread morphology of these cells that could play an important role in their trophic and scavenger functions. On the other hand, the incompleteness of the reconstitution of the bone resorption and of tissue macrophage densities in many tissues, including spleen, adrenal, large intestine, kidney medullar, adult liver, and peritoneal cavity, as well as the hematologic abnormalities in Csf1op/Csf1op; TgCS5/+ mice, indicates that the secreted CSF-1 isoforms have important roles.
Specific aspects of the different roles of csCSF-1 and secreted CSF-1 in the maintenance of normal tissue macrophage densities, in the regulation of normal hematopoiesis, and in supporting the full function of osteoclasts bear further comment. Tissues normally possessing high macrophage densities, which were not fully restored in Csf1op/Csf1op mice by TgCS expression, generally have a rich blood supply. These tissues, which include spleen, liver, adrenal gland, and kidney would have good access to the secreted CSF-1 isoforms found in the circulation and may therefore be preferentially dependent on circulating CSF-1 for their maintenance. Indeed, we have previously shown that restoration of circulating CSF-1 in Csf1op/Csf1op mice by daily injection of rhCSF-1 efficiently restores the macrophage densities of liver, kidney, and spleen (including MOMA-1+ cells).55 These results are consistent with the failure of TgCS to reconstitute macrophage densities in these tissues, and together they indicate the importance of regulation by the secreted isoforms. For example, newborn Csf1op/Csf1op mouse liver has a nearly wild-type macrophage density due to the transplacental transfer of CSF-1.70 However, by 3 months of age, their macrophage density drops to approximately one third the wild-type density55,70 (Table 3). Correction of the adult macrophage density in Csf1op/Csf1op mice was observed by restoration of circulating CSF-155 but not by expression of csCSF-1 (Table 3). In contrast, tissues normally possessing lower densities of resident macrophages, which in Csf1op/Csf1op mice were restored completely by TgCS expression, for example, synovial membrane and dermis, generally have relatively poor blood circulation. csCSF-1 on local CSF-1-producing cells was sufficient to maintain their macrophage populations, which at best were only partially restored in Csf1op/Csf1op mice injected with rhCSF-1.55
TRAP+ osteoclasts in Csf1op/Csf1op; TgCS/+ mice were similar in number to their wild-type controls and possessed a similar overall morphology. However, the obvious increase in trabecular bone in the subepiphyseal region of femurs of Csf1op/Csf1op; TgCS5/+ mice (Figure 4) indicated that there is a very significant delay in the progress of trabecular bone resorption. Given the apparently normal osteoclastogenesis, there are at least 2 possible explanations for the delayed bone resorption in the Csf1op/Csf1op; TgCS/+ mice. First, csCSF-1 may be not able to support full osteoclastic bone resorption activity. This could be due to the failure of csCSF-1 to appropriately activate the required signaling pathways. Differential signaling has been shown for soluble and cell surface stem cell factor.71,72 Alternatively, in the absence of soluble CSF-1, csCSF-1 may not be able to support the migration of osteoclasts from one resorption site to another. Second, by using transgenic mice expressing the lac Z reporter gene driven by CSF-1 promoter/first intron,32 we have shown that major CSF-1-producing cells in long bone reside in trabecular bone, especially near the junction of the hypertrophic chondrocytes and trabecular bone in the epiphyseal region (Figure 4). Because soluble CSF-1 has been shown to be chemotactic for macrophages,73,74 the diffusible, secreted glycoprotein and proteoglycan CSF-1 produced in this area may be able to act as a morphogen to form a growth factor gradient that attracts osteoclasts toward the subepiphyseal regions of the bone. Establishing the relative contribution of csCSF-1 and soluble CSF-1 to osteoclastic bone resorption will require further study.
Beside the direct effect of CSF-1 on mononuclear phagocytic lineage development, it also plays an important role in general hematopoiesis through the CSF-1 requirement for osteoclastogenesis and bone marrow cavity development in early postnatal life.75,76 In addition, we and others have shown that early hematopoietic progenitor cells express the CSF-1R,77-79 suggesting a role of CSF-1 in early hematopoiesis. TgCS was not able to fully reconstitute the hematopoiesis in the Csf1op/Csf1op; TgCS/+ mice. A significant component of this phenotype may be due to the residual osteopetrosis resulting from the delayed bone resorption. However, compared with Csf1op/+ mice, the decrease in granulocytes in the bone marrow and spleens of Csf1op/Csf1op; TgCS/+ mice and their increase in Csf1op/Csf1op mice (Table 4) suggest that csCSF-1 may have an inhibitory effect on granulocytopoiesis.
A variety of growth factors have been found to have membrane-anchored isoforms with specific biologic roles.80-82 A juxtacrine stimulation model was proposed in which membrane-bound growth factor-mediated cell adhesion through growth factor receptor binding participates in biologic processes, such as the homing of cells to tissue locations.83 The interaction of membrane-spanning cytokines with their receptors on adjacent cells has been shown to support cellular survival and differentiation, notably for c-kit ligand in mice84-87 and boss in Drosophila.88
Binding of soluble CSF-1 to the CSF-1R triggers serial molecular events, including the dimerization and autophosphorylation of CSF-1R, as well as activation of the tyrosine kinase activity of the CSF-1R and the tyrosine phosphorylation of downstream molecules. The CSF-1/CSF-1R complex then undergoes clathrin-dependent endocytosis. The internalized ligand/receptor complex is subsequently degraded in the lysosome.89-91 Internalization and degradation of the CSF-1R serves to down-regulate the soluble CSF-1 cellular signaling. Because receptor-mediated internalization of the membrane-anchored ligand/receptor complex does occur in the case of the Drosophila boss protein,92 an important question is whether the csCSF-1/CSF-1R complex undergoes a similar internalization. Furthermore, because both quantitative and qualitative differences between c-kit-mediated signals activated by soluble and membrane-bound stem cell factor have been demonstrated,71,72 it is important to know whether similar differences exist between csCSF-1/CSF-1R-mediated signaling and signaling by soluble CSF-1 and if so, how does csCSF-1/CSF-1R signaling affect cell differentiation, function, and migration? Further investigation of these questions is required to fully understand the in vivo role of csCSF-1 and to interpret the phenotypes we have observed in the Csf1op/Csf1op; TgCS/+ mice. However, even without the answers to these mechanistic questions, Csf1op/Csf1op; TgCS/+ mice are a useful model system for the studies of the role of CSF-1 in a variety of pathologic processes.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-08-2739.
Supported by National Institutes of Health grant CA32551 (E.R.S.), the Albert Einstein College of Medicine Cancer Center grant 5P30-CA13330, an American Society of Hematology Fellow Scholar Award (X.-M.D.), and a Leukemia and Lymphoma Society Special Fellow Award (X.-M.D.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Zoya Aronova for technical assistance and members of the AECOM transgenic, FACS, histopathology, and analytical imaging facilities for assistance in different aspects of the work. We acknowledge kind gifts of the CSF-1 promoter clone from Dr Michael Cole, MOMA-1 antibody from Dr G. Kraal, and monoclonal antibody F4/80 from Dr David Hume.