Abstract
Contemporary treatment of pediatric acute lymphoblastic leukemia (ALL) requires the assignment of patients to specific risk groups. We have recently demonstrated that expression profiling of leukemic blasts can accurately identify the known prognostic subtypes of ALL, including T-cell lineage ALL (T-ALL), E2A-PBX1, TEL-AML1, MLL rearrangements, BCR-ABL, and hyperdiploid karyotypes with more than 50 chromosomes. As the next step toward developing this methodology into a frontline diagnostic tool, we have now analyzed leukemic blasts from 132 diagnostic samples using higher density oligonucleotide arrays that allow the interrogation of most of the identified genes in the human genome. Nearly 60% of the newly identified subtype discriminating genes are novel markers not identified in our previous study, and thus should provide new insights into the altered biology underlying these leukemias. Moreover, a proportion of the newly selected genes are highly ranked as class discriminators, and when incorporated into class-predicting algorithms resulted in an overall diagnostic accuracy of 97%. The performance of an array containing the identified discriminating genes should now be assessed in frontline clinical trials in order to determine the accuracy, practicality, and cost effectiveness of this methodology in the clinical setting.
Introduction
Pediatric acute lymphoblastic leukemia (ALL) is a heterogeneous disease with subtypes that differ markedly in their cellular and molecular characteristics as well as their response to therapy and subsequent risk of relapse.1,2 Contemporary treatment protocols achieve overall long-term survival rates of 70% to 80% by tailoring the intensity of therapy to a patient's risk of relapse.1,3-6 Therefore, accurate assignment of individual patients to risk groups is a critical issue for optimal outcome.
Current risk assignment incorporates clinical characteristics (age, sex), basic laboratory studies (presenting white blood cell count and presence or absence of leukemia in cerebral spinal fluid), as well as characteristics of the leukemic blasts (immunophenotype, cytogenetics, molecular diagnostics for the presence of translocation-encoded fusion transcripts, and response to therapy).7,8 The early risk features were primarily identified from epidemiologic studies correlating clinical characteristics with outcome data. For example, infants were found to have poorer overall survival than children between the ages of 2 and 10 years old.9 Immunophenotypic characterization of leukemic blasts subsequently revealed that patients with T-cell lineage ALL (T-ALL) had a higher risk for relapse than B-precursor ALL.10 More recently, detailed cytogenetic analysis coupled with the subsequent molecular cloning of the underlying lesions have resulted in the identification of genetically distinct subtypes of B-lineage ALL. These include t(9;22)(BCRABL), t(1;19)(E2A-PBX1), t(12;21)(TEL-AML1), rearrangement in the MLL gene on chromosome 11q23, and hyperdiploid karyotype with more than 50 chromosomes.1,2,11,12
Completion of the human genome project and the development of high throughput parallel expression analysis using DNA-based microarrays has allowed the use of this information in cancer classification.13-15 We have previously demonstrated that expression profiles obtained using oligonucleotide microarrays can be used to accurately identify 6 prognostic subtypes of pediatric ALL.16 In that study, gene expression profiles used in combination with supervised learning algorithms had an overall diagnostic accuracy of 96%, a level comparable with current multidisciplinary diagnostic techniques. Thus, the single platform of microarray analysis may be able to achieve a risk assignment accuracy equal to that currently obtained using the collective expertise of multiple laboratories.
As a next step toward development of a custom diagnostic microarray, the current study used the Affymetrix HG-U133 set of microarrays to identify diagnostic discriminating genes from a larger proportion of the human genome. This 2-chip set provides an almost 3-fold increase in the number of genes evaluated compared with our original study, interrogating an estimated 39 000 transcripts. From our previous study of 327 pediatric ALL cases, we chose 132 representative cases to evaluate using the HG-U133A and B microarrays. The data obtained provide direct support for use of microarray-based expression profiling as a single platform for the diagnosis of the known prognostic subtypes of pediatricALL. Moreover, the data provide additional insights into the biology underlying the clinical differences between these leukemia subgroups.
Materials and methods
Gene expression profiling
The preparation of mononuclear cell suspensions from diagnostic bone marrow aspirates, extraction of total RNA, and preparation of hybridization solutions has been previously described (detailed protocols are available in the Supplemental Information from our original paper at http://www.stjuderesearch.org/data/ALL1).16 All protocols and consent forms were approved by the hospital's institutional review board, and informed consent was obtained from parents, guardians, or patients (as appropriate). Individual hybridization solutions from our previous study had been stored at –80°C since initial hybridization (approximately 1 year). These solutions were thawed and hybridized to Affymetrix HG-U133A and HG-U133B oligonucleotide microarrays (Affymetrix, Santa Clara, CA) according to Affymetrix protocols. In 2 cases where the original hybridization solutions were no longer available, replicate viably frozen mononuclear cell preparations from the diagnostic bone marrow aspirate were obtained, RNA isolated, cDNA and cRNA synthesized, labeled, fragmented, and hybridized as previously described.16
After sample hybridization, arrays were then stained with phycoerythrinconjugated streptavidin (Molecular Probes, Eugene, OR). Antibody amplification was performed with biotinylated antistreptavidin (Vector Laboratories, Burlingame, CA), followed by staining with phycoerythrin-conjugated streptavidin (Molecular Probes). Arrays were scanned using a laser confocal scanner (Agilent, Palo Alto, CA) and then analyzed with Affymetrix Microarray suite 5.0 (MAS 5.0). Detection values (present, marginal, or absent) were determined by default parameters, and signal values were scaled by global methods to a target value of 500. Microarray scan images were visually inspected for apparent defects, and Affymetrix internal controls were used to monitor the success of hybridization, washing, and staining procedures. Minimal quality control parameters for inclusion in the study included more than 10% present calls and a glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) 3′/5′ ratio of 3 or less. The arrays included in this study had an average percent present call of 35.9% for the A chip and 21.0% for the B chip (combined average of 28.5%).
Statistical analysis
The dataset was separated into a training set (n = 100) and test set (n = 32). Prior to analysis, a variation filter was applied to remove any probe set that showed minimal variation across the dataset, and thus contributed minimally, if at all, to the discrimination of leukemia subtypes. Specifically, probe sets were eliminated from further analysis if the number of cases with a present call was less than one half the number of samples comprising the leukemia subgroup under analysis (in the parallel format as described below the number 5 was used since this represents approximately one half of the number of BCR-ABL leukemia samples, the smallest group in the training set), had a signal value less than 100 in all samples in the dataset, or had a maximal signal value in the dataset – minimal signal value in the dataset that was less than 100. In addition, all signal values with absent or marginal calls were reset to 1, while probe sets with a present “P” call and a signal less than 100 had the signal reset to 100. The values for signals from the Affymetrix control sets were removed prior to analysis.
The identification of subtype-discriminating probe sets was performed exclusively using cases in the training set and was approached using both a parallel and a differential diagnosis decision tree format. In the parallel approach, class-discriminating probe sets were first identified by defining probe sets that had an expression pattern that was specific to one class compared with all other cases in the training set. The top 20 to 50 class-discriminating probe sets for each class were then combined and used in supervising learning algorithms to assign cases to 1 of 7 classes (T-ALL, E2A-PBX1, MLL rearrangement, BCR-ABL, TEL-AML1, hyperdiploid with more than 50 chromosomes, or other). As previously described,16 in the differential diagnosis decision tree format, class-discriminating genes were first selected for T-ALL against all other cases. The T-ALL cases were then removed, the variation filter reapplied, and then class-discriminating genes were selected for E2A-PBX1 cases. The latter cases were then removed and the process repeated proceeding sequentially through TEL-AML1, BCR-ABL, MLL rearrangement, and hyperdiploid with more than 50 chromosomes.
The class-discriminating genes were primarily selected using a chi-square metric. In this procedure, an entropy-based discretization method was first applied to identify genes whose expression across the dataset showed differentiation between class and nonclass.17 The assigned discretized value for the gene was then used in a chi-square calculation to determine if the association with a class was more than would be expected by random chance. The stronger the association with the class, the larger the chi-square value calculated. For the genes that could not be discretized, their chi-square values were set to zero. To evaluate the statistical significance of the discriminating genes, we used a permutation test in which for each class, case labels were randomly reassigned to generate new groups of identical size. The label permutated data were discretized again and the chi-square values were recalculated. The permutation test was repeated for a total of 1000 times. The true chi-square values for each probe set were then compared with the values generated from the 1000 permutations to determine how many times a chi-square value for a probe set in a randomly labeled group was greater than that obtained for the true class distinction. A P value was calculated as the number of times the chi-square value exceeded the true value in the 1000 permutations.
The identified discriminating genes were then used in either unsupervised clustering algorithms (2-dimensional hierarchic clustering or principal component analysis [PCA], GeneMaths software version 1.5; Applied Maths, Austin, TX), or supervised learning algorithms to build classifiers that could identify the specific genetic subgroup. The supervised learning algorithms used included k-nearest neighbors (k-NN), support vector machine (SVM), and an artificial neural network (ANN).16,18-20 Performance of each model was initially assessed by 3-fold cross-validation on a randomly selected stratified training set. True error rates of the best performing classifiers were then determined using the remaining one fourth of the samples as a blinded test group. Class assignment required that a sample's calculated node value exceed a statistically determined confidence level in order for it to be assigned to a class. Details of the supervised learning algorithms and their use are described on the Blood website; see the Supplemental Document link at the top of the online article.
Gene comparison
Due to multiple design advances used to produce the HG-U133 microarrays, most probe sets differ between the HG-U95Av2 and HG-U133 series microarrays. Affymetrix has provided comparison spreadsheets to assist in comparing data obtained on HG-U95Av2 with that obtained with HGU133A and B array sets. These comparison spreadsheets provide a mechanism for finding a “best match” or “good match” between array sets as determined by probe set sequence identity. However, individual genes may be represented by multiple probe sets that share little or no sequence homology. Using these tables underestimates the information in common between U95 and U133 experiments. Comparisons were therefore made based on Unigene reference for each probe set as supplied by Affymetrix. However, many of the probe sets are annotated as simply expressed sequence tags (ESTs). To further define these sites, target and/or consensus sequences were used to search public databases.
Supplemental document
Additional information on the samples, methods, statistical analysis, and results from the comparison of microarray gene expression levels with mRNA levels determined by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) are available in the Supplemental Document. The primary data are available at http://www.stjuderesearch.org/data/ALL3.
Results
The Affymetrix HG-U133 set of microarrays allows the interrogation of an estimated 39 000 transcripts, thus providing an almost 3-fold increase in the number of genes evaluated compared with the HG-U95Av2 microarray used in our original study.16 To determine if the additional expression data provided by these microarrays would both enhance our ability to accurately diagnose and subclassify pediatric ALL, and provide additional insights into the underlying biology of the different genetic subtypes of ALL, we selected a subset of our original 327 diagnostic pediatric ALL samples to reanalyze using these higher density microarrays. Case selection was based on providing a representation of the known prognostic ALL subtypes including t(9;22)(BCR-ABL), t(1;19)(E2APBX1), t(12;21)(TEL-AML1), rearrangement in the MLL gene on chromosome 11q23, and hyperdiploid karyotype with more than 50 chromosomes. Since our goal was to define expression profiles that could be used to accurately diagnose the known prognostic subtypes of ALL, we chose to overrepresent these subtypes compared with what is normally seen in a random population of childhood leukemia patients. A total of 132 samples met these criteria and had sufficient material remaining to be used for this analysis. The list of samples and the subtype distribution of the cases used in this study are as shown in Table 1, and Tables S1 and S2 in the Supplemental Document.
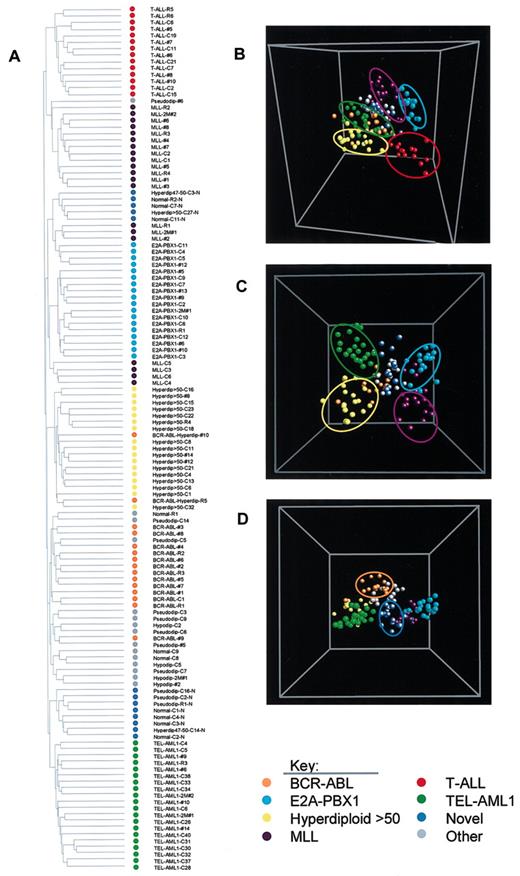
After the application of the variation filter to the 132 diagnostic leukemia samples, 26 825 probe sets from combined U133A and B microarrays remained. In an initial analysis of these data, we used 2 complementary unsupervised clustering algorithms— 2-dimensional hierarchic clustering and principle component analysis (PCA)—to assess the major subgroupings of the leukemia cases based solely on the expression profiles of these genes across the entire dataset of 132 cases. In agreement with our previous study,16 these unbiased clustering algorithms demonstrated that the pediatric ALL cases cluster primarily into 7 major subtypes: T-ALL and 6 subtypes of B-cell lineage ALL corresponding to (1) rearrangement in the MLL gene on chromosome 11q23, (2) t(1;19)(E2A-PBX1), (3) hyperdiploid with more than 50 chromosomes, (4) t(9;22)(BCR-ABL), (5) the previously described novel subgroup,16 and (6) t(12;21)(TEL-AML1) (Figure 1). In addition, a heterogeneous group of B-lineage cases was identified that lacked any of the defined genetic lesions and failed to cluster into the novel subgroup (the primary data are available at http://www.stjuderesearch.org/data/ALL3). Several of these leukemia subtypes formed distinct branches when all differentially expressed genes were used in the 2-dimensional hierarchic clustering algorithm (T-ALL, hyperdiploid with more than 50 chromosomes, and TEL-AML1), whereas other subtypes clustered in multiple branches, suggestive of gene expression differences within these subclasses (Figure 1A). Using PCA, the distinct nature of the B-cell lineage subtypes is better appreciated when the T-ALL cases were removed from the analysis (compare Figure 1B, which includes the T-ALL cases, to the 2 views of the analysis after the removal of the T-ALL cases shown in Figure 1C-D). It should be noted that 100% diagnostic accuracy appears to be achieved for only 2 of the leukemia subtypes (T-ALL and TEL-AML1). This finding is not unexpected when using an unsupervised clustering algorithm that calculates similarities and differences based on the complete set of expressed probe sets. If relatively few probe sets distinguish a specific genetic subtype of leukemia, then the calculated distances used by the algorithms will not reflect the influence of these important discriminating probe sets. These observations indicate the need to use supervised learning algorithms to achieve optimal diagnostic accuracy by gene expression profiling.
Distinct leukemia subtypes can be defined based exclusively on their expression profiles. Expression profiles were obtained on leukemic blasts from 132 diagnostic bone marrow aspirates and the data analyzed using (A) an unsupervised 2-dimensional clustering algorithm and (B-D) principle component analysis (PCA). In this analysis the cases in the training and test sets were combined, and the analysis was performed with the 26 825 genes from the Affymetrix U133A and B microarrays that varied in their expression across this dataset. (A) A 2-dimensional hierarchic clustering was performed using Pearson correlation coefficient and unweighted pair group method using arithmetic averages. (B) Multidimensional scaling plot of all cases using PCA. (C) Multidimensional scaling plot of B-lineage ALL cases (n = 118). (D) The identical multidimensional scaling plot as shown in panel C except the plot was rotated 90°. Each case is represented by a sphere and is color coded to indicate the genetic subgroup to which it belongs: BCR-ABL (orange), E2A-PBX1 (aqua), hyperdiploid with more than 50 chromosomes (yellow), MLL (purple), T-ALL (red), TEL-AML1 (green), novel cases (blue), and unclassified cases (gray).
Distinct leukemia subtypes can be defined based exclusively on their expression profiles. Expression profiles were obtained on leukemic blasts from 132 diagnostic bone marrow aspirates and the data analyzed using (A) an unsupervised 2-dimensional clustering algorithm and (B-D) principle component analysis (PCA). In this analysis the cases in the training and test sets were combined, and the analysis was performed with the 26 825 genes from the Affymetrix U133A and B microarrays that varied in their expression across this dataset. (A) A 2-dimensional hierarchic clustering was performed using Pearson correlation coefficient and unweighted pair group method using arithmetic averages. (B) Multidimensional scaling plot of all cases using PCA. (C) Multidimensional scaling plot of B-lineage ALL cases (n = 118). (D) The identical multidimensional scaling plot as shown in panel C except the plot was rotated 90°. Each case is represented by a sphere and is color coded to indicate the genetic subgroup to which it belongs: BCR-ABL (orange), E2A-PBX1 (aqua), hyperdiploid with more than 50 chromosomes (yellow), MLL (purple), T-ALL (red), TEL-AML1 (green), novel cases (blue), and unclassified cases (gray).
As shown in Figure 1, the previously defined novel ALL subgroup was easily identified using these higher density microarrays. This subtype of ALL clustered into 2 distinct branches by 2-dimensional hierarchic clustering and into a single cluster by PCA. Since the goal of this paper is to define the expression profile of the known prognostic ALL subtypes, we chose not to further analyze the novel subgroup, since the clinical and biologic importance of this group remains to be defined.
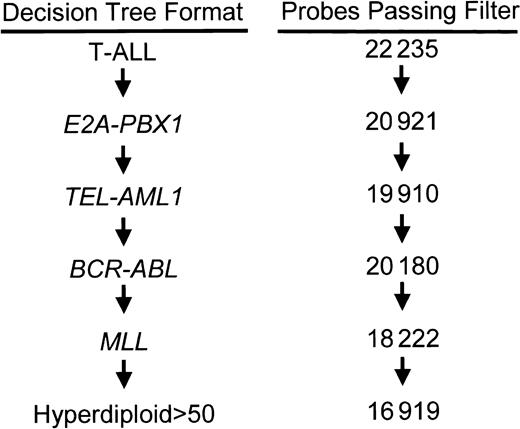
We next used statistical methods to identify probe sets that were the best discriminators of the individual leukemia subtypes. Class distinction can be approached using a parallel decision format, in which sets of genes are selected that can discriminate each individual leukemia subtype against all other cases. Alternatively, class assignment can be approached using a differential decision tree approach (Figure 2), as previously described.16,21 Since the latter method provided the highest level of accuracy in our original study, we chose to use this approach for most of our analysis. Briefly, in selecting genes for use in the differential diagnosis decision tree approach, we first select genes that discriminate T-ALL from all other cases. The T-ALL cases were then removed, the filter reapplied, and then discriminating genes selected for E2A-PBX1. The E2A-PBX1 cases were then removed and the process repeated sequentially as illustrated in Figure 2. Cases not assigned to one of these classes are left unassigned. The use of this decision tree format directly influences the selection of genes, allowing the selection of discriminating genes for groups lower down the tree that might also be expressed by subtypes higher in the tree. For purposes of comparison, the lists of probe sets selected using both the decision tree and parallel formats are presented in the Supplemental Document.
Structure of the differential diagnostic decision tree. Gene discovery and class prediction were performed following the illustrated decision tree. At each level, the dataset was filtered to remove genes that showed minimal variation in their level of expression between the genetic subtype (class) under evaluation and all subtypes that fall below it in the decision tree (nonclass). Numbers at the right represent the number of probe sets that pass the variance filter at each level. The cases assigned to a diagnostic subgroup are removed prior to progressing to the next level in the algorithm. Cases that pass through the entire decision tree without being assigned to a class are classified as “other.”
Structure of the differential diagnostic decision tree. Gene discovery and class prediction were performed following the illustrated decision tree. At each level, the dataset was filtered to remove genes that showed minimal variation in their level of expression between the genetic subtype (class) under evaluation and all subtypes that fall below it in the decision tree (nonclass). Numbers at the right represent the number of probe sets that pass the variance filter at each level. The cases assigned to a diagnostic subgroup are removed prior to progressing to the next level in the algorithm. Cases that pass through the entire decision tree without being assigned to a class are classified as “other.”
Discriminating genes were selected using a chi-square metric on the 100 cases in the training set. The number of discriminating probe sets per leukemia subtype at a statistical significance level of P values less than or equal to .001 (as determined by a permutation test) were T-ALL, 2063; E2A-PBX1, 1059; TEL-AML1, 805; BCR-ABL, 201; MLL chimeric genes, 726; and hyperdiploid with more than 50 chromosomes, 994. The marked differences in number of discriminating genes for the various leukemia subtypes suggest that significant differences exist in the global gene expression profiles of the leukemias that are likely to be dependent on the specificdefining genetic lesions. The number of discriminating genes per leukemia subtype was nearly identical when calculated using a parallel format in which each leukemia subtype was compared against all other leukemias (data not shown and see Supplemental Document), demonstrating that the observed differences are intrinsic to the leukemia, and not significantly influenced by the method used to compare and contrast the individual leukemia subtypes.
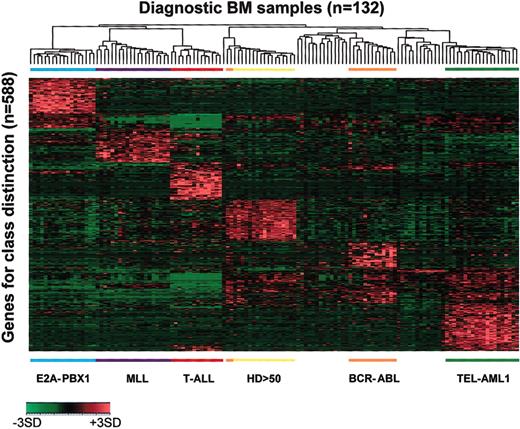
The expression profiles obtained using the top 100 ranked probe sets for the 6 prognostically important subgroups are illustrated in Figure 3 using the 2-dimensional hierarchic clustering algorithm (the lists of discriminating genes are contained in the Supplemental Document, Tables S3-S14). In this figure, each column corresponds to a single leukemia sample and each row represents the expression level of a probe set across the cases, with red representing expression above, and green below the mean. Of the 600 discriminating probe sets selected using the decision tree format, 12 were in the top 100 for more than one group and were therefore used only once in the figure, resulting in a total of 588 discriminating probe sets. As multiple probe sets for the same gene are present on Affymetrix microarrays, the top 100 ranked probe sets represent between 75 and 92 distinct genes, depending on the leukemia subtype (Table 2 and Supplemental Document). As shown, distinct groups of either overexpressed or underexpressed genes distinguish cases defined by BCR-ABL, E2A-PBX1, hyperdiploid with more than 50 chromosomes, MLL gene rearrangement, T-ALL, and TEL-AML1.
Expression profile of pediatric ALL diagnostic bone marrow blasts. Shown is a 2-dimensional hierarchic cluster of 132 pediatric ALL diagnostic bone marrow samples (columns) versus the top 100 chi-square ranked probe sets (rows) for each of the 6 diagnostic subgroups of ALL. There were 12 probe sets identified as useful in discriminating more than one class and they are represented only once in the diagram. Probe set signal values are normalized to the mean for the dataset, and values for each individual case are represented by a color, with red representing deviation above the mean and green representing deviation below the mean. Genetic subtypes are indicated across the bottom of the panel.
Expression profile of pediatric ALL diagnostic bone marrow blasts. Shown is a 2-dimensional hierarchic cluster of 132 pediatric ALL diagnostic bone marrow samples (columns) versus the top 100 chi-square ranked probe sets (rows) for each of the 6 diagnostic subgroups of ALL. There were 12 probe sets identified as useful in discriminating more than one class and they are represented only once in the diagram. Probe set signal values are normalized to the mean for the dataset, and values for each individual case are represented by a color, with red representing deviation above the mean and green representing deviation below the mean. Genetic subtypes are indicated across the bottom of the panel.
A comparison was undertaken to determine how many of the top 100 discriminating probe sets selected using the U133 data represent new class-discriminating genes that were not previously identified using the U95Av2 microarray.16 Of the top 100 probe sets selected using the new platform, 40% were genes that had previously been determined to be class discriminators using the U95Av2 microarray (Table 2 and Supplemental Document, Tables S3-S8). Moreover, 90% of the top 100 discriminating probe sets selected using the U95Av2 microarray were identified as genes in the U133 data that had statistically significant chi-square value (P < .01 by permutation test, data not shown) confirming them as class discriminators; however, many of these previously selected genes were ranked below the top 100 for the U133 arrays, as would be expected if better discriminating genes were present on the U133 microarrays. Of the remaining U133 selected discriminating genes, 40% had no representation on the U95Av2 platform, and thus represent new class predictors (Table 2). The remaining 20% were represented on the HG-U95Av2 microarray but had not been selected as class discriminators in the previous study (Table 2). Further examination of these latter genes demonstrated that in the U95Av2 data, some of these genes showed a trend in their pattern of expression that suggested they might be class discriminators; however, the class association was below a statistical significance level (P < .01 by a permutation test, data not shown). It thus appears that the improved oligonucleotide design in the U133, coupled with the use of signal amplification, results in an increase in the sensitivity/specificity of detection of these discriminating genes. This interpretation was confirmed for a subset of these genes using quantitative RT-PCR assays that demonstrated their differential expression across the leukemia subtypes (see Supplemental Document, Table S18). Thus, almost 60% of the top 100 ranked genes selected using the U133 platform represent new leukemia subtype discriminating genes (Table 2).
Biologic insights from the new class-defining genes
Interestingly, the overall quantitative pattern of expression of discriminating genes varied significantly between leukemia subtypes (Table 3). Regardless of whether using genes selected through a parallel or decision tree format, the fold increase of class-discriminating genes was significantly higher for leukemia subtypes defined by chimeric transcription factors than for those with BCR-ABL or hyperdiploid with more than 50 chromosomes. These data suggest that the quantitative global changes in a cell's expression profile vary markedly depending on the genetic lesion(s) that underlie the initiation of the leukemic process. Moreover, in hyperdiploid with more than 50 chromosomes the observation that class-discriminating genes had an average fold increase of only 2 suggests that the enhanced expression of these genes may be the result of a simple increase in gene dosage secondary to chromosomal trisomies.
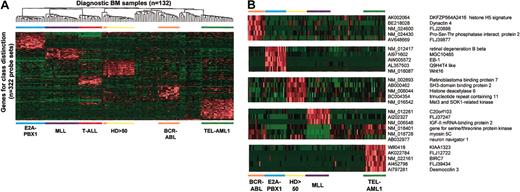
The expression patterns of the newly identified genes across the entire dataset are illustrated in Figure 4A. As shown, the new genes effectively cluster the individual subtypes of pediatric ALL. Shown in Figure 4B are examples of new genes whose expression is limited to a single B-cell lineage class, and therefore function not only as class discriminators in the decision tree format, but are also class discriminators in a parallel format in which a class is distinguished against all others. Thus, these genes have the potential of serving as unique class-specific diagnostic or therapeutic targets. In addition, these genes may provide unique insights into the underlying biology of the different leukemia subtypes. For example, among the new discriminating genes in E2A-PBX1–expressing leukemias were 2 genes, EB-1 and Wnt16, that had previously been shown by more conventional methods to be overexpressed in this leukemia subtype (Supplemental Document, Table S4).22,23 In addition, the retinal degeneration B beta gene24 and a number of novel ESTs were identified as being uniquely overexpressed in this leukemia subtype (Figure 4B), whereas the SOCS2-negative regulators of cytokine signaling were found to be underexpressed (Supplemental Document, Table S4).25
New class-discriminating genes. (A) Shown is a 2-dimensional hierarchic cluster of ALL cases using the newly identified class discriminators from the U133A and B microarrays. Probe set signal values are normalized to the mean for the dataset and then for each individual case are represented by color, with red representing deviation above the mean and green representing deviation below the mean. Genetic subtypes are indicated across the bottom of the panel. (B) Shown are selected genes highly correlated with individual genetic subtypes of ALL. Probe set signal values are normalized to the mean for the dataset, and values for each individual case are represented by color, with red representing deviation above the mean and green representing deviation below the mean. The leukemia subtype is indicated at the bottom of the figure. GenBank accession numbers and gene symbols or DNA sequence names are listed on the right side of the panel.
New class-discriminating genes. (A) Shown is a 2-dimensional hierarchic cluster of ALL cases using the newly identified class discriminators from the U133A and B microarrays. Probe set signal values are normalized to the mean for the dataset and then for each individual case are represented by color, with red representing deviation above the mean and green representing deviation below the mean. Genetic subtypes are indicated across the bottom of the panel. (B) Shown are selected genes highly correlated with individual genetic subtypes of ALL. Probe set signal values are normalized to the mean for the dataset, and values for each individual case are represented by color, with red representing deviation above the mean and green representing deviation below the mean. The leukemia subtype is indicated at the bottom of the figure. GenBank accession numbers and gene symbols or DNA sequence names are listed on the right side of the panel.
Other newly identified discriminating genes of potential interest that were identified in TEL-AML1 leukemias included a putative gene, KIAA1323, localized to chromosome 18q11.1 that encodes a 795–amino acid protein that has 8 ankyrin repeat domains and a C-terminal RING finger domain. This combination of domains is identified in only a limited number of mammalian proteins, most notably BRCA-1–associated RING domain 1 (BARD1), a regulator of the breast cancer 1, early onset (BRCA1) tumor suppressor.26 Other genes overexpressed in the subtype include desmocollin,27 FLJ12722, which encodes a novel protein of unknown function, and a member of the IAP family of apoptosis inhibitors, BIRC7, which is overexpressed 25-fold.28
Expression profiling accurately identifies the prognostic subtypes of pediatric ALL
A major goal of this study was to determine the accuracy of identifying prognostically important ALL genetic subtypes by expression profiling. To assess this, the top 50 class-discriminating genes identified using a chi-square metric were used in an ANN-based supervised learning algorithm (the highest ranked probe set for each gene was used in this analysis). Class assignment used the decision tree differential diagnostic format described in Figure 2 and required that the node value for assignment exceed a statistically defined confidence level (Supplemental Document). As shown in Table 4, using this approach resulted in exceptionally accurate class prediction in a randomly selected training set that consisted of three fourths of the total cases (100 cases). When this classification model was then applied to a blinded test set consisting of the remaining 32 samples, an overall accuracy of 97% was achieved for class assignment (Table 4 and Supplemental Document). To control for overfitting of the data, we performed 10 additional rounds of this analysis where for each round new training and test sets were developed; genes were then reselected using the new training set, and then their performance was assessed on the new test set. This resulted in an average accuracy of class assignment in the blinded test sets of 97.2%, with a range from 93.8% to 100% (Supplemental Document, Tables S15 and S16). Although the number of genes required for optimal class assignment varied between classes, the overall diagnostic accuracy was essentially the same using either the top 20 or top 50 genes per class (Supplemental Document, Tables S15 and S16). A similar level of accuracy was achieved using a variety of other supervised learning algorithms, including κ-NN and SVM (Supplemental Document, Table S17), and using these algorithms in a parallel approach (data not shown).
Interestingly, of the rare misclassification errors, 2 were cases of BCR-ABL–expressing ALL that by gene expression analysis were classified as hyperdiploid with more than 50 chromosomes. The karyotype of these cases showed the presence of both the Philadelphia chromosome and a hyperdiploid karyotype consisting of more than 50 chromosomes—including trisomy of chromosomes X and 21 (data not shown). The expression profile thus correctly identified the presence of the hyperdiploid with more than 50 chromosomes class; however, since each case is assigned to only a single class, the algorithm failed to correctly identify the presence of BCR-ABL. Nevertheless, the data presented demonstrate the exceptional accuracy of this single platform for the diagnosis of the prognostically important subtypes of pediatric ALL.
Discussion
The recent application of gene expression profiling to the classification of human hematopoietic malignancies has shown great promise.13,14,16,29-31 In aggregate, these studies have not only raised the possibility of achieving a more accurate diagnosis of clinically relevant disease subtypes, but have also suggested that a single standardized diagnostic platform may be obtainable. With an expanding representation of the entire human genome being placed on microarrays, it should in the near future be possible to identify the entire genome-wide expression profile of a leukemic cell. This undoubtedly will lead to both a greater understanding of the altered biology underlying the formation of the leukemic clone, as well as an enhancement in our ability to identify leukemia subtype–specific expression signatures. The hope is that some of the identified class-specific molecules will in turn prove to be useful therapeutic targets. As a next step to developing a leukemia diagnostic microarray, we have expanded on our previous analysis of the expression profile of pediatric ALL by now assessing the expression of approximately 33 000 genes, representing a substantial proportion of the human genome, in 132 diagnostic bone marrow samples.
Using a chi-squared metric followed by permutation test to identify genes significantly associated with a particular class, we selected discriminating genes for each of the 6 major prognostic subgroups, T-ALL, E2A-PBX1, TEL-AML1, BCR-ABL, MLL rearrangement, and hyperdiploid with more than 50 chromosomes. Approximately 90% of the original top 100 ranked discriminating genes identified using the U95Av2 array were reselected as discriminating genes using this new platform, validating these genes as useful class discriminators. Of the top 100 genes selected using this new platform, approximately 60% of the genes were not selected using the U95Av2 array, and thus are new class discriminators. Importantly, a proportion of the newly selected genes is highly ranked as class discriminators, and thus incorporating these into a class-predicting algorithm should result in a further increase in the diagnostic accuracy of gene expression profiling. Consistent with this prediction, when the top 50 genes identified using the U133 arrays per class were used in an ANN supervised learning algorithm on a true test set consisting of 32 samples, an overall prediction accuracy of 97% was achieved. Comparable accuracies were achieved using the discriminating genes in other supervised learning algorithms. Thus, the identified discriminating genes provide exceptional diagnostic accuracies on this idealized set of pediatric ALL samples.
What level of diagnostic accuracy will be needed to move this approach into a clinical setting? How many genes will be needed per leukemia subtype to achieve the required accuracy, and what computational approach will be necessary? Moreover, will expression profiling be able to accurately identify specific leukemia subtypes on diagnostic bone marrow aspirates containing less than 75% blasts, a not infrequent occurrence? Lastly, will the method prove to be cost effective? Although each of these questions will need to be directly addressed before expression profiling can move into a frontline diagnostic setting, we can start to formulate answers to some of these questions based on the data presented in this paper.
Although 100% accuracy is desired, the reality is that our presently used diagnostic approaches frequently fall far short of this goal. Identification of some of the ALL subtypes can be based solely on cytogenetic analysis, a method whose accuracy is primarily dependent on the level of expertise of the practitioner. Depending on the institution, the detection of an abnormal karyotype in ALL varies between 75% and 90%32,33 —a variance that is indicative of errors. Moreover, although some efforts have been expended to standardize the molecular-based assays used for the detection of translocation-encoded fusion transcripts,34 many institutions still rely on home-brew assays with variable performance. In addition, for MLL gene rearrangements RT-PCR, genomic Southern blots, and fluorescence in situ hybridization–based assays have all proved of utility; however, no single assay provides complete diagnostic accuracy.35-37 In this setting a standardized single platform for class assignment could serve not only to improve diagnostic accuracy, but also to facilitate cross comparisons between therapeutic protocols.
The data presented suggest that expression profiling using the U133 microarrays should provide a level of accuracy that is comparable, if not superior, to that obtained using standard diagnostic methodologies. However, our data also suggest that a platform that contains only probes specific for the detection of normal genes may be inadequate to attain 100% diagnostic accuracy. Specifically, the diagnosis of BCR-ABL–expressing leukemia remains problematic, in part because of the close relationship between the expression profiles of BCR-ABL and hyperdiploid with more than 50 chromosomes ALLs. Moreover, rare cases of BCR-ABL have a hyperdiploid karyotype. Since these 2 leukemia subtypes sit at opposite extremes of the spectrum of relative risk of relapse, this misdiagnosis could have serious clinical consequences. One possible approach to enhance the performance of expression profiling in accurately diagnosing BCR-ABL might be to incorporate oligonucleotide probes specific for this chimeric transcript into the microarray. Since this translocation results in only a limited variety of chimeric transcripts,38 the sequences are readily available and could be easily incorporated into a custom diagnostic microarray. This modification would allow the rapid and accurate identification of a BCR-ABL fusion transcript within a sample. A similar approach could also be designed for the other translocationencoded chimeric transcripts, possibly further enhancing the accuracy with which we can diagnose these subtypes.
The defined expression profiles are likely to vary in their ability to accurately subclassify leukemia in a bone marrow aspirate that is composed of less than 75% blasts. This is in part due to the marked differences seen in the magnitude of gene expression changes between subtypes. The high expression of class-specific genes in T-ALL, E2A-PBX1, TEL-AML1, and MLL rearrangement should provide a relatively sensitive approach for accurately subclassifying leukemias. By contrast, the magnitude of the expression signatures of the other leukemia subtypes makes it unlikely that they could accurately subclassify leukemia in a sample containing a minority of leukemic cells. Although the exact level of diagnostic sensitivity will need to be experimentally determined, simple methods exist for enriching the percentage of leukemic blasts, thus making it unlikely that this would be a serious concern in the diagnostic setting.
The newly identified subtype-specific genes should provide further insights into the altered biology that underlies the abnormal growth of these leukemias. Although a number of potentially important genes have been highlighted, detailed analysis of each leukemic subtype will be required to maximize the insights that can be gained from this new dataset. It is important to realize that key insights are likely to come not only from class-specific genes, but also from the analysis of genes involved in key cellular regulatory pathways, regardless of whether they are uniquely expressed in a particular leukemia subtype. Also it will be important to compare gene expression patterns between leukemic cells and their normal cellular counterpart, from which they are believed to arise. For B-cell lineage ALL these latter cells are believed to be normal bone marrow–derived B cells. Experiments are in progress to identify the expression profile of normal bone marrow–derived B cells and the differences that exist between these normal profiles and those that characterize the different subtypes of B-lineage ALL.
In summary, our results suggest that expression profiling should provide a single standardized platform for the accurate diagnosis of the major prognostic subtypes of pediatric ALL. Moreover, the database generated through this study should provide a unique resource for the investigation of the altered biology that underlies the various genetic subtypes of acute leukemia.
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2003-01-0338.
Supported in part by National Cancer Institute grants P01 CA71907-06 (J.R.D.), CA-21765 (Cancer Center CORE grant to SJCRH), T32-CA70089, and by the American Lebanese and Syrian Associated Charities (ALSAC) of SJCRH.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank the staff of the Molecular Pathology Laboratory, the Hartwell Center for Bioinformatics and Biotechnology, and the members of the Hematological Malignancies program at St. Jude Children's Research Hospital (SJCRH).