Abstract
Delivery of biologically active peptides into human polymorphonuclear neutrophils (PMNs) has implications for studying cellular functions and may be therapeutically relevant. The transcription factor nuclear factor-κB (NF-κB) regulates the expression of multiple genes controlling inflammation, proliferation, and cell survival. PMNs play a crucial role in first-line defense. Targeting NF-κB in these cells may promote apoptosis and therefore facilitate resolution of inflammation. We used an 11-amino acid sequence NEMO-binding domain (NBD) that selectively inhibits the IKKγ (NEMO)/IKKβ interaction, preventing NF-κB activation. An HIV-TAT sequence served as a highly effective transducing shuttle. We show that lipopolysaccharide (LPS), granulocyte-macrophage colony-stimulating factor (GM-CSF), and dexamethasone (DEX) significantly reduced apoptosis after 20 hours. LPS, but not GM-CSF or DEX, activated NF-κB as shown by IκBα degradation, NF-κB DNA binding, and transcriptional activity. The TAT-NBD blocked LPS-induced NF-κB activation and NF-κB–dependent gene expression. TAT-NBD accelerated constitutive PMN apoptosis dose dependently and abrogated LPS-delayed apoptosis. These results provide a proof of principle for peptide delivery by TAT-derived protein transduction domains to specifically inhibit NF-κB activity in PMNs. This strategy may help in controlling various cellular functions even in short-lived, transfection-resistant primary human cells.
Introduction
The introduction of biologically active peptides into human polymorphonuclear neutrophils (PMNs) may help in clarifying intracellular signal transduction and ultimately could have therapeutic implications. However, all peptide delivery methods available thus far are inefficient. Previously, peptide transduction domains (PTDs) were identified that shuttle even large proteins in excess of 100 000 Da into mammalian cells in vitro and in vivo.1 These “Trojan-horse” peptides include the homeodomain of Antennapedia (a Drosophila transcription factor), a short amino acid sequence of HIV-1, and the herpes simplex virus 1 (HSV-1) structural protein VP22.2-4 We explored the use of an 11-amino acid sequence (amino acids 47-57) from the HIV TAT protein (HIV-TAT) in PMNs to target nuclear factor-κB (NF-κB).
NF-κB is a transcription factor controlling gene expression during inflammation, immunity, cell proliferation, stress response, and apoptosis.5-8 NF-κB is activated by many agents including cytokines, viral infection, UV radiation, and free radicals.9 In unstimulated cells, NF-κB is sequestered in the cytoplasm by tightly bound inhibitors (IκBα, IκBβ, IκBϵ). The inhibitors are phosphorylated and rapidly degraded, allowing NF-κB to translocate into the nucleus and activate target genes. IκBα is phosphorylated on serine residues by the multicomponent IκB kinase (IKK) containing 2 catalytic subunits (IKKα and IKKβ) and one regulatory subunit (IKKγ).10-13 In contrast to other cell types, the role of NF-κB in PMNs is incompletely characterized due to rapid NF-κB degradation by proteolytic enzymes, difficulties with PMN transfection, and the lack of specific NF-κB inhibitors. Previous studies with pharmacologic compounds, such as pyrrolidine dithiocarbamate (PDTC), SN50, and gliotoxin, have suggested that NF-κB is involved in regulating PMN apoptosis.14-16 However, the specificity of these agents has been questioned.17-19 We used a highly specific small peptide to block the interaction of IKKγ with the IκB kinase complex (IKK).20 We generated a linear 2-domain peptide containing the NEMO-binding domain (NBD) to specifically block NF-κB and the TAT-PTD to shuttle NBD into PMNs.
Materials and methods
Materials
Granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin 8 (IL-8) were obtained from R&D Systems (Wiesbaden-Nordenstedt, Germany). Lipopolysaccharide (LPS), tumor necrosis factor α (TNF-α), dexamethasone (DEX), Ficoll-Hypaque, and propidium iodide were from Sigma (Deisenhofen, Germany). Dextran was from Amersham Pharmacia (Amsterdam, The Netherlands), the polyclonal rabbit antibodies against IκBα (C-21), p65 (A), Rel-B, c-Rel, and Jun-B were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); p50 and p52 antibodies were from Rockland and Upstate Biotechnology (Lake Placid, NY). Monoclonal IKKα antibody (FL-418) was obtained from Pharmingen (Heidelberg, Germany). Horseradish peroxidase–labeled donkey antirabbit immunoglobulin G (IgG) was from Amersham (Braunschweig, Germany); Hanks balanced salt solution (HBSS), phosphate-buffered saline (PBS), RPMI 1640, and trypan blue were from Biochrom (Berlin, Germany); and the ApoAlert annexin V apoptosis kit was from Clontech (Palo Alto, CA). Fetal calf serum (FCS) was purchased from Gibco (Karlsruhe, Germany). Endotoxin-free reagents and plastic disposables were used in all experiments.
Isolation of human neutrophils
PMNs from healthy human donors were isolated from heparinized whole blood by red blood cell sedimentation with dextran 1%, followed by Ficoll-Hypaque density gradient centrifugation. Erythrocytes were lysed by incubation with hypotonic saline for 15 seconds. PMNs were spun down (1050 rpm, 10 minutes) and reconstituted in HBSS with calcium and magnesium or, when cultured in RPMI 1640 supplemented with 10% FCS, l-glutamine, penicillin, and streptomycin. Samples were incubated in polypropylene tubes and kept at 37°C in 5% CO2. The cell viability was detected in every cell preparation by trypan blue exclusion and found to be greater than 99%. The percentage of PMNs in the suspension was more than 95% by Wright-Giemsa staining and by light microscopy. Informed consent was obtained according to the Declaration of Helsinki, and the research protocol was approved by the Franz Volhard Clinic Ethics Committee.
Peptide synthesis
We synthesized several peptides encompassing the HIV TAT-PTD (amino acids 47-57). The TAT-PTD was synthesized together with an 11-amino acid sequence representing the IKKβ NEMO-binding domain (TALD-WSWLQTE) or a mutant peptide with Trp→Ala mutations (TAL-DASALQTE), respectively. The wild-type peptide inhibits the IKKβ/IKKγ (NEMO) interaction and has been termed NBD. Both were extensively characterized by May et al.20 For transduction efficacy studies, the TAT-PTD was synthesized with an NH2-terminal fluorescein isothiocyanate (FITC) via a 6-aminohexancarbonic acid (Ahx) linker. Peptide synthesis was performed by Biosynthan (Berlin, Germany) using a PSSM-8 from Shimadzu (Duisburg, Germany) and the final product was analyzed by ion exchange chromatography (fast protein liquid chromatography [FPLC]), followed by measuring the mass spectrum in a time of flight (TOF) analyzer (Shimadzu). Increasing concentrations of FITC or TAT-FITC (50 nM to 1 μM) were added to 5 × 105 cells in 100 μL PBS. Samples were incubated for 10 minutes at 37°C, washed in 500 μL ice-cold PBS, and analyzed by flow cytometry. PMNs were read using a fluorescence-activated cell sorter (FACscan) (Becton Dickinson, Heidelberg, Germany) and 10 000 events per sample were collected in listmode using Lysis II software for data acquisition and analysis. In a different set of experiments, cells were treated with 1 μM FITC, FITC-TAT-NBD, and FITC-TAT-Ctrl (MUT) under the aforementioned conditions. Finally, 5 μM of either FITC-TAT-NBD or FITC-TAT-Ctrl was added to 500 μL heparinized whole blood. After 20 minutes of incubation at 37°C, erythrocytes were removed by hypotonic lysis, and cells were washed in 500 μL PBS. Cells were assayed using flow cytometry with a gate set on neutrophils according to light scatter characteristics. FITC staining of neutrophils was analyzed in the FL1-channel.
Confocal microscopy
To analyze the introduction of the mutant peptide and the NBD peptide, isolated PMNs or whole blood were incubated for 20 minutes with buffer control, 5 μM free FITC, FITC-TAT-NBD, and FITC-TAT-Ctrl, respectively. Unfixed live cells were washed once and incubated for another 15 minutes in the live cell DNA dye DRAQ5. DNA dye was added after red blood cell lysis when whole blood was used. Confocal microscopy was performed on a Zeiss LSM510 using ×40 NA 1.2 C-Apochromat water immersion objective as well as ×63 and ×100 NA 1.4 Planapochromat oil immersion objectives. FITC was excited with a 488 nm argon laser line and detected using a high-frequency transduction (HFT) 488-nm beam splitter and 505-nm long-pass emission filter. Laser power and detector settings were identical between the different samples. The reproducibility of the results was verified by 2 separate investigators.
Western blot
Samples were incubated for 5 minutes at 95°C in loading buffer (250 mM Tris [tris(hydroxymethyl)aminomethane]–HCL, pH 6.8, with 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.01% bromphenol blue, 10% β-mercaptoethanol). Five to 20 μg protein was loaded per lane, electrophoresed on a 10% SDS-polyacrylamide gel, and transferred to a nitrocellulose membrane. The membranes were blocked with triethanolamine-buffered saline (TBS-Tween)/5% skim milk for 1 hour, and incubated overnight with an antibody to IκBα (1:1000 dilution). Membranes were washed and incubated with a secondary antibody (horseradish peroxidase–labeled donkey antirabbit IgG, 1:2500). The blot was developed by incubation in a chemiluminescence substrate (enhanced chemiluminescence [ECL], Amersham) and exposed to an x-ray film. Equal loading of protein was confirmed by stripping and reprobing the blots for total p38. Densitometry of IκBα was performed with scanned x-ray films and the National Institutes of Health (NIH) image program.
Cytoplasmic and nuclear extract preparation
Cytoplasmic and nuclear extracts were prepared according to McDonald et al21 and Dignam with several modifications.22 The reaction was stopped by adding an equal amount of ice-cold RPMI supplemented with diisopropylfluorophosphate (DFP; 2 mM final concentration). Cells were centrifuged at 2000g for 2 minutes at 4°C and pellets were resuspended with a hypotonic buffer A (HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] 10 mM, pH 7.5, KCl 10 mM, EGTA [ethylene glycol tetraacetic acid] 0.1 mM, EDTA [ethylenediaminetetraacetic acid] 0.1 mM, pH 8.0) containing an antiprotease cocktail (2 mM DFP, 1 mM AEBSF [aminoethylbenzene sulfonyl fluoride], 1 mM PMSF [phenylmethylsulfonyl fluoride], 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin A, 0.5 mM benzamidine, 1 mM DTT [dithiothreitol]). Cells were kept on ice for 10 minutes, followed by the addition of NP40 (0.1% final concentration). Cells were vortexed briefly and pelleted. The resulting supernatant was referred to as the cytoplasmic fraction and stored at –80°C. For the preparation of nuclear extracts, we observed the best results when omitting NP40. Apparently NP40 itself led to the degradation of the nuclei. After the addition of buffer A, cells were vortexed and spun down at 1000g (10 minutes, 4°C) to pellet the nuclei and to remove intracellular granules that may cause nuclear degradation. The pellet was washed again with buffer A, and the hypertonic, high-salt buffer C was added (HEPES 20 mM, pH 7.5, NaCl 0.4 M, EDTA 1 mM, EGTA 1 mM, glycerol 20%) together with the protease inhibitor cocktail. After centrifugation (10 minutes at 14 000g, 4°C), the supernatant was collected and referred to as the nuclear fraction. Protein measurements were done using the Bio-Rad assay (Bio-Rad, Munich, Germany).
To minimize degradation of PMNs an alternative method was also used. Nuclear extracts were prepared using nitrogen cavitation by a modified method of McDonald et al.21 Briefly, PMNs (3 × 107) were stimulated, and activation was stopped as described (see “Cytoplasmic and nuclear extract preparation”). Centrifuged cell pellets were resuspended in relaxation buffer and pressurized with constant stirring under a N2 atmosphere (350 psi, 15 minutes, 4°C) in a nitrogen bomb. Cavitates were spun down at 1000g (10 minutes, 4°C) to pellet most of the nuclei, and the resulting supernatants were recentrifuged to pellet the remaining nuclei. Both nuclear pellets were combined and washed in relaxation buffer. Pellets were resuspended again, NaCl was then added to 400 mM final concentration, and incubated on ice for 20 minutes prior to centrifugation (14 000g, 10 minutes, 4°C). The resulting supernatant was referred to as the nuclear fraction.
Electrophoretic mobility shift assay
For the electrophoretic mobility shift assay (EMSA), nuclear extracts (5 μg protein) were incubated with 20 000 cpm of a 22-bp oligonucleotide containing the NF-κB consensus sequence that had been labeled with [α-32] adenosine triphosphate (ATP) T4 polynucleotide kinase. Incubations were performed for 30 minutes at room temperature in the presence of poly deiodinase/deoxycytidine (poly/dI-dC) and 20 mM HEPES, containing 60 mM KCl, 4% Ficoll, 5 mM DTT, and 0.5 μg/μL nuclease-free bovine serum albumin (BSA). Probes were subjected to electrophoresis on native 5% polyacrylamide gels and autoradiographed. For supershift assays the indicated antibody was added 15 minutes before the addition of the radiolabeled probe. To determine the specificity of shifted bands, excess unlabeled oligonucleotide or a mutated cold probe was added to the nuclear extracts 10 minutes before addition of the radiolabeled probe. For the mutated cold probe 2 amino acids of the oligonucleotide (H2K) were replaced and hybridization of 2 complementary single oligonucleotides was performed. The oligonucleotides for H2K were forward primer: 5′ GATCCAGGGCTGGGGATTCCCCATCTCCACAGG3′, reverse primer: 5′ GATCCCTGTGGAGATGGGGAATCCCCAGCCCTG 3′; and for the mutated form, forward primer: 5′ GATCCAGGGCTAGCGATTCCCCATCTCCACAGG 3′.
Equal excess of the oligonucleotides was used for competition experiments.
IκB kinase activity assay
To assay IκB kinase activity, 6 to 8 × 106 cells were stimulated with 1 μg/mL LPS or 20 ng/mL GM-CSF or left untreated after pretreatment with TAT-NBD or the mutant control. Cell extracts were essentially prepared as described previously with several modifications.23 Whole cell lysates were prepared using lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 10% glycerol) with the protease inhibitor cocktail as described. Cell equivalents were used for immunoprecipitation, which was performed in lysis buffer. Before the addition of 1 μg monoclonal IKKα antibody (Pharmingen) extracts were precleared with protein A-Sepharose (Amersham) for 30 minutes at 4°C. Binding of the antibody was carried out overnight at 4°C, and the next day 25 μL protein A-Sepharose was added for additionally 2 hours. The protein A-Sepharose was washed 3 times with lysis buffer and one time with kinase buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, 20 mMATP, 20 μM β-glycerophosphate, 1 mM DTT). Then 15 μL kinase buffer including purified recombinant IκBα and 3 μCi (0.111 MBq) [γ-32]ATP were added to the protein A-Sepharose immunocomplex and the kinase reaction was incubated for 20 minutes at 37°C. Samples were boiled and subjected to 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by autoradiography. Equal loading was confirmed by IKKα Western blot experiments.
Quantitative RT-PCR
Total RNAs were isolated according to a Qiagen protocol including DNase treatment. Quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed using TaqMan technology (Applied Biosystems, Weiterstadt, Germany). Reverse transcription was carried out according to the Superscript protocol (Invitrogen, De Schelp, The Netherlands). TaqMan RT-PCR was performed using the Master Mix (Applied Biosystems). The quantification was checked for each sample using probes for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Primers and probes were designed using the primer express program (Applied Biosystems). The following oligonucleotides were used for IκBα: forward primer 5′ CCCTGTAATGGCCGGACTG 3′, reverse primer 5′ AGGAGTGACACCAGGTCAGGA 3′ and the probe Fam 5′ CCTTCACCTC GCAGTGGACCTGC 3′ Tamra. RT-PCR and quantification were performed using the TaqMan 5700 (Applied Biosystems). For quantification of the amount of RNA present in the various samples, the fluorescence signal was measured at each PCR cycle and the increase in the fluorescence normalized reporter signal (RN) documented in an amplification plot. Using nontemplate controls, the threshold was set in the log phase to subtract unspecific fluorescence signals. Cycle threshold (ct) values were determined for each sample. In short, the ct value difference (Δct) was used to calculate the factor of differential expression (2Δct). Results were imported in an Excel spreadsheet and analyzed according to the standard curve method.
Assessment of apoptosis
Flow cytometry was used to measure DNA content in ethanol-permeabilized cells at the single-cell level as described previously.24 The assessment of apoptosis with propidium iodide takes advantage of the fact that activated endonucleases generate low-molecular-weight DNA fragments in apoptotic cells. After membrane permeabilization with ethanol, these fragments leak out, resulting in decreased DNA-content in apoptotic cells, while DNA content in nonapoptotic cells remains unchanged. Propidium iodide stains DNA of permeabilized cells, and normal PMNs can be appreciated by a G0/1 peak using flow cytometry. In contrast, apoptotic cells with reduced DNA are reflected by a sub-G0/1 peak. The percent of apoptotic cells was determined by the separation between the sub-G0/1 apoptotic cells and the G0/1 nonapoptotic population. Briefly, freshly isolated or cultured cells were spun at 200g for 7 minutes at 4°C and resuspended in PBS containing 0.5 mM EDTA. Chilled 95% ethanol was added to a final concentration of 70% and the cells were stored at –20°C overnight. PMNs were pelleted and resuspended in 100 μL PBS/0.5 mM EDTA/1% BSA. Then, 100 μL PBS containing 200 μg DNase-free RNase and 400 μL PBS containing 50 μg propidium iodide were added. After 6 to 8 hours in the staining mixture at 4°C, PMNs were analyzed using a FACscan and 10 000 events per sample were collected in listmode using Lysis II software for data acquisition and analysis. For annexin V staining, freshly isolated or cultured cells (5 × 105) were washed with PBS and pelleted, followed by resuspension in 200 μL binding buffer. Then 10 μLof annexin V was added to the cells. After incubation in the dark at room temperature for 10 minutes, the cells were subjected to flow cytometry analysis. Annexin V binds only to the surface of apoptotic cells and the percentage of annexin V+ cells by flow cytometry reflects the apoptotic population.
Statistical analysis
Results are given as mean ± SEM. Comparisons between 2 groups were done using paired Wilcoxon rank tests. Comparisons between multiple groups were done using Kruskal Wallis tests. Specific differences between multiple groups were then determined by use of a Bonferroni post hoc test on the ranked values.
Results
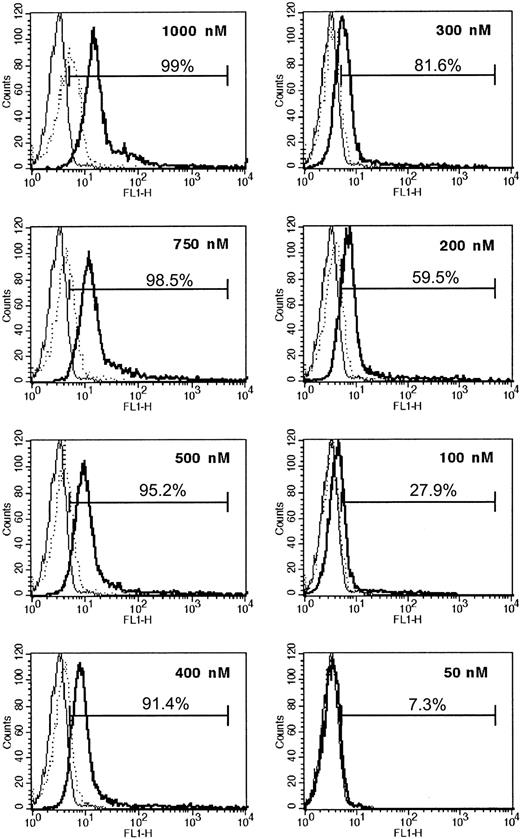
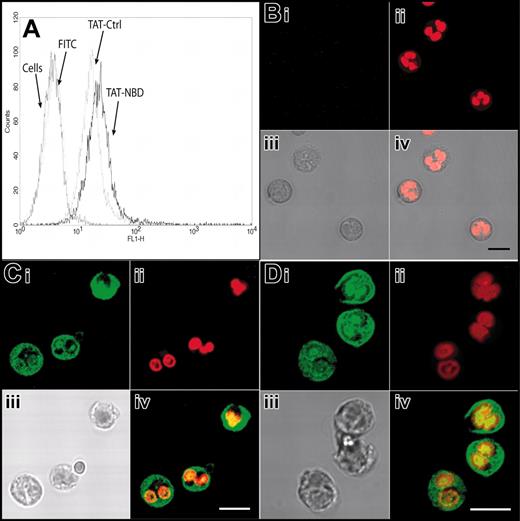
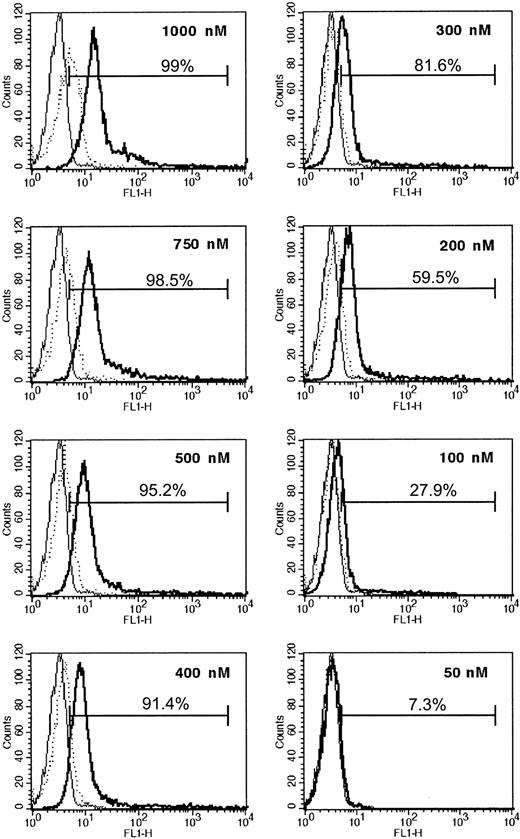
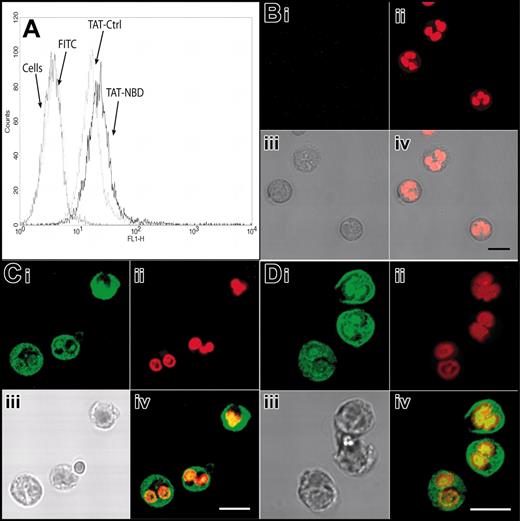
We first explored the possibility that the PTD of the HIV-TAT peptide (amino acids 47-57) can be effectively transduced into PMNs. We linked FITC to the HIV-TAT PTD. By flow cytometry, we found that free FITC did not enter PMNs, whereas FITC linked to the TAT sequence rapidly stained PMNs in a dose-dependent manner (Figure 1). FITC-TAT, 1 μM, resulted in staining of almost 100% of the cells. To exclude the possibility that the increased staining of TAT-FITC–incubated PMNs seen in the flow cytometry assay was merely a result of molecules sticking to the outer cell surface, we performed confocal microscopy. TAT-FITC was transduced into the cell, whereas no intracellular fluorescence was observed when cells were incubated with free FITC molecules (data not shown). Next, we assessed the transduction of FITC-labeled TAT-NBD and the mutant TAT-Ctrl into isolated neutrophils as well as into neutrophils with the incubation step carried out in whole blood. We performed flow cytometry and confocal microscopy (Figure 2A-D). Figure 2A demonstrates that FITC-TAT-NBD and FITC-TAT-Ctrl (mutant form) stained almost 100% of the cells, whereas no staining was observed with free FITC. Confocal microscopy shows that TAT-NBD and TAT-Ctrl (Figure 2C-D) localized throughout the cytoplasm and the nucleus, whereas free FITC (Figure 2B) was not transduced into cells. TAT peptides are taken up into the cytoplasm and nucleus and accumulated to a lower extent in chromatin-dense areas. In addition, FITC-labeled TAT peptides were transduced into the entire granulocyte population even when added to whole blood (data not shown). These results indicate that molecules containing an NH2-terminal TAT-PTD can efficiently enter human PMNs.
Dose-dependent introduction of TAT-FITC into PMNs. PMN staining after incubation with free FITC or TAT-FITC was studied by flow cytometry. Cells were treated for 10 minutes with increasing concentrations of free FITC or TAT-FITC as indicated. After washing, the fluorescence intensity was assessed in the FL-1 channel. Thin solid lines represent cell autofluorescence; dotted lines, fluorescence after free FITC incubation; and bold lines, FL-1 staining of cells incubated with TAT-FITC. The data indicate increased staining of PMNs that were incubated with TAT-FITC, but not with free FITC.
Dose-dependent introduction of TAT-FITC into PMNs. PMN staining after incubation with free FITC or TAT-FITC was studied by flow cytometry. Cells were treated for 10 minutes with increasing concentrations of free FITC or TAT-FITC as indicated. After washing, the fluorescence intensity was assessed in the FL-1 channel. Thin solid lines represent cell autofluorescence; dotted lines, fluorescence after free FITC incubation; and bold lines, FL-1 staining of cells incubated with TAT-FITC. The data indicate increased staining of PMNs that were incubated with TAT-FITC, but not with free FITC.
Transduction of TAT-FITC assayed by flow cytometry and confocal microscopy. Transduction of TAT-NBD and TAT-Ctrl peptides was studied by flow cytometry (A) and confocal microscopy (B-D). (A) Isolated neutrophils were incubated with buffer, free FITC, FITC-labeled TAT-Ctrl, or FITC-labeled TAT-NBD (5 μm) for 20 minutes at 37°C. Flow cytometry shows that both the FITC-labeled TAT-NBD and the TAT-Ctrl peptides stained almost 100% of the cells. For confocal microscopy, live cells were treated with the indicated peptides and control FITC (green; Bi,Ci,Di), washed once, and incubated in the live-cell DNA dye DRAQ5 (red; Bii,Cii,Dii). Samples were immediately analyzed by confocal microscopy without fixation. The images demonstrate that TAT-NBD (C) and TAT-Ctrl (D) localize throughout the cytoplasm and the nucleus, whereas free FITC (B) does not transduce into cells. In each case, an overlay image is shown at the bottom right-hand side (Biv,Civ,Div). To better visualize the localization of the TAT-Ctrl and TAT-NBD, only the green and red channels were overlaid. TAT peptides are taken up into the cytoplasm and nucleus and accumulate to a lower extent in chromatin dense areas. Scale bars correspond to 50 μm in the upper panels (i-ii) and 10 μm in the lower panels (iii-iv).
Transduction of TAT-FITC assayed by flow cytometry and confocal microscopy. Transduction of TAT-NBD and TAT-Ctrl peptides was studied by flow cytometry (A) and confocal microscopy (B-D). (A) Isolated neutrophils were incubated with buffer, free FITC, FITC-labeled TAT-Ctrl, or FITC-labeled TAT-NBD (5 μm) for 20 minutes at 37°C. Flow cytometry shows that both the FITC-labeled TAT-NBD and the TAT-Ctrl peptides stained almost 100% of the cells. For confocal microscopy, live cells were treated with the indicated peptides and control FITC (green; Bi,Ci,Di), washed once, and incubated in the live-cell DNA dye DRAQ5 (red; Bii,Cii,Dii). Samples were immediately analyzed by confocal microscopy without fixation. The images demonstrate that TAT-NBD (C) and TAT-Ctrl (D) localize throughout the cytoplasm and the nucleus, whereas free FITC (B) does not transduce into cells. In each case, an overlay image is shown at the bottom right-hand side (Biv,Civ,Div). To better visualize the localization of the TAT-Ctrl and TAT-NBD, only the green and red channels were overlaid. TAT peptides are taken up into the cytoplasm and nucleus and accumulate to a lower extent in chromatin dense areas. Scale bars correspond to 50 μm in the upper panels (i-ii) and 10 μm in the lower panels (iii-iv).
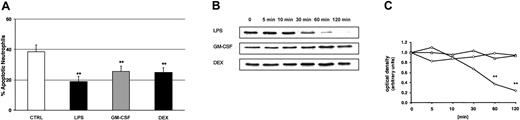
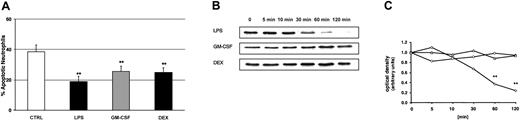
We used LPS, GM-CSF, and DEX to study PMN apoptosis. Cells were cultured overnight for 20 hours in the presence of buffer control, 100 ng/mL LPS, 20 ng/mL GM-CSF, or 10–7 M DEX, respectively. Flow cytometry with propidium iodide staining confirmed that all 3 compounds significantly delayed apoptotic cell death (Figure 3A). Constitutive apoptosis was 38% ± 4%. LPS decreased this number to 19% ± 3%, GM-CSF to 26% ± 3%, and DEX to 25% ± 3%.
Delay of PMN apoptosis by LPS, GM-CSF, and DEX, and LPS-induced degradation of IκBα. (A) Samples were incubated with buffer control, 100 ng/mL LPS, 20 ng/mL GM-CSF, and 10–7 M DEX. Cells were harvested after 20 hours, permeabilized in ethanol, stained with propidium iodide, and assayed by flow cytometry. These experiments indicate significantly delayed apoptosis by LPS, GM-CSF, and DEX (n = 6, **P < .01). The values represent mean ± SEM. (B) The effect of 100 ng/mL LPS, 20 ng/mL GM-CSF, and 10–7 M DEX on IκBα degradation was studied by Western blot (n = 3). PMNs were harvested at the indicated time points; cytoplasmic extracts were prepared and analyzed for IκBα. (C) For densitometric analysis of the IκBα expression, the optical density value of the first band (time point 0 minutes) was set 1.0 and the following time points represent the relative changes of IκBα intensity. Only LPS led to a significant degradation of IκBα after 60 and 120 minutes (**P < .01). ⋄ indicates LPS; ○, GM-CSF; and ▵, DEX.
Delay of PMN apoptosis by LPS, GM-CSF, and DEX, and LPS-induced degradation of IκBα. (A) Samples were incubated with buffer control, 100 ng/mL LPS, 20 ng/mL GM-CSF, and 10–7 M DEX. Cells were harvested after 20 hours, permeabilized in ethanol, stained with propidium iodide, and assayed by flow cytometry. These experiments indicate significantly delayed apoptosis by LPS, GM-CSF, and DEX (n = 6, **P < .01). The values represent mean ± SEM. (B) The effect of 100 ng/mL LPS, 20 ng/mL GM-CSF, and 10–7 M DEX on IκBα degradation was studied by Western blot (n = 3). PMNs were harvested at the indicated time points; cytoplasmic extracts were prepared and analyzed for IκBα. (C) For densitometric analysis of the IκBα expression, the optical density value of the first band (time point 0 minutes) was set 1.0 and the following time points represent the relative changes of IκBα intensity. Only LPS led to a significant degradation of IκBα after 60 and 120 minutes (**P < .01). ⋄ indicates LPS; ○, GM-CSF; and ▵, DEX.
To document the importance of NF-κB in delaying apoptosis, we next determined whether or not the NF-κB pathway was activated by LPS, GM-CSF, or DEX. We first studied NF-κB activity by assaying IκBα degradation and nuclear NF-κB DNA-binding activity. PMNs were treated for up to 120 minutes with LPS, GM-CSF, and DEX, respectively. By Western blot, we show that incubation of PMNs with LPS, but not with GM-CSF or DEX, resulted in degradation of IκBα (Figure 3B). The kinetics of NF-κB activation stimulated by LPS revealed that the first effect occurred after 30 minutes and increased up to 120 minutes. GM-CSF and DEX did not lead to any measurable IκBα degradation. Figure 3C shows the corresponding densitometric analysis from 3 parallel experiments. Statistical analysis confirmed that LPS-induced degradation of IκBα was significant after 60 and 120 minutes.
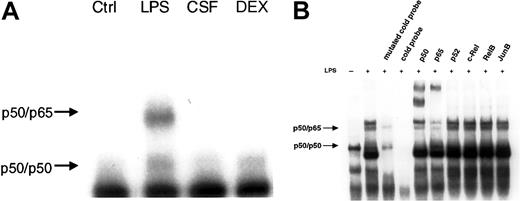
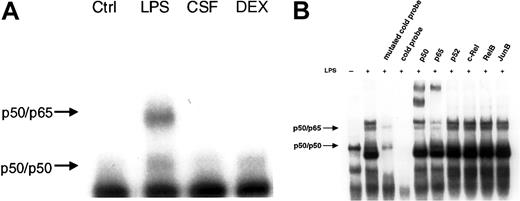
We then investigated the effect of LPS, GM-CSF, and DEX on NF-κB activity by EMSA. The results demonstrate NF-κB activation only in response to LPS (Figure 4A). GM-CSF and DEX did not induce NF-κB activation nor did they reduce any basal activity. Abrogated basal NF-κB activity by DEX was reported in murine lymphocytes earlier.25 We also performed supershift experiments with antibodies directed against each of the 5 known mammalian members of the NF-κB family to determine which subunits of the NF-κB family participated in the observed complexes (Figure 4B). An antibody against p50 strongly reduced the upper and lower band with the formation of an intensive supershifted complex. Serum against p65 caused a decrease of the upper band with the appearance of a supershifted complex, whereas no change in the intensity of bands was observed with anti-p52, –c-Rel, -RelB, and -JunB, an irrelevant antibody. From these results, the lower band can be identified as a p50/p50 homodimer and the upper band as the p50/p65 heterodimer. Specificity was verified by competition with a 20-fold excess of an unlabeled NF-κB probe (cold probe), and, additionally, with a mutated cold probe (Figure 4B).
LPS, but not GM-CSF or DEX, led to NF-κB–binding activity. (A) PMNs were either left untreated or treated with 1 μg/mL LPS, 20 ng/mL GM-CSF (CSF), or 10–7 M DEX for 60 minutes; nuclear extracts were prepared and analyzed by EMSA using an H2K-binding site probe for NF-κB (n = 3). Only LPS increased NF-κB–binding activity in PMNs. The amount of nuclear extract used for the binding reaction was 5 μg protein for all samples. (B) Specificity of the bands was assessed by competition with a cold probe versus a mutated cold probe and supershift experiments. A 20-fold excess of unlabeled NF-κB probe and mutated cold probe and 5 specific antibodies against the family of the different members of the NF-κB family and one irrelevant antibody were added to nuclear extracts before incubation with labeled NF-κB oligonucleotide probe (n = 3). Two specific bands could be identified: the lower band as the p50/p50 homodimer and the upper band as the p50/p65 heterodimer.
LPS, but not GM-CSF or DEX, led to NF-κB–binding activity. (A) PMNs were either left untreated or treated with 1 μg/mL LPS, 20 ng/mL GM-CSF (CSF), or 10–7 M DEX for 60 minutes; nuclear extracts were prepared and analyzed by EMSA using an H2K-binding site probe for NF-κB (n = 3). Only LPS increased NF-κB–binding activity in PMNs. The amount of nuclear extract used for the binding reaction was 5 μg protein for all samples. (B) Specificity of the bands was assessed by competition with a cold probe versus a mutated cold probe and supershift experiments. A 20-fold excess of unlabeled NF-κB probe and mutated cold probe and 5 specific antibodies against the family of the different members of the NF-κB family and one irrelevant antibody were added to nuclear extracts before incubation with labeled NF-κB oligonucleotide probe (n = 3). Two specific bands could be identified: the lower band as the p50/p50 homodimer and the upper band as the p50/p65 heterodimer.
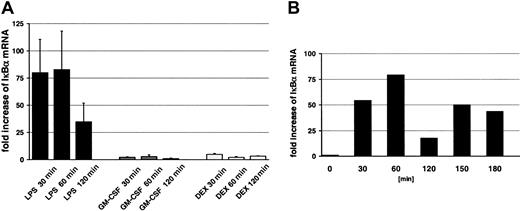
Finally, we used quantitative RT-PCR to study the effect of LPS, GM-CSF, and DEX on activation of NF-κB–dependent gene expression. IκBα, the cytoplasmic inhibitor of NF-κB, is itself generated in a NF-κB–dependent manner and was selected for that reason. LPS resulted in an approximately 80-fold increase of IκBα mRNA, whereas only marginal effects were observed when cells were incubated with GM-CSF or DEX (Figure 5A). Figure 5A also demonstrates that the LPS-induced IκBα mRNA expression decreased rapidly after its peak. Therefore, by RT-PCR we performed a kinetic profile with prolonged LPS stimulation up to 180 minutes. A representative figure is depicted in Figure 5B (n = 3). We observed in all experiments an oscillatory kinetic pattern with 30- to 60-minute time intervals, whereas the time of occurrence of the first peak of transcriptional activity differed between the donors, reflecting well-known interindividual variability in PMN responses.
Relevant up-regulation of IκBα mRNA by LPS and expression of oscillatory kinetics under prolonged stimulation. (A) PMNs were incubated with 1 μg/mL LPS, 20 ng/mL GM-CSF (CSF), or 10–7 M DEX for the indicated time points and synthesis of IκBα mRNA was assessed by quantitative RT-PCR (n = 6 for LPS; n = 3 for GM-CSF and DEX). Total RNAs were isolated according to a Qiagen protocol including DNase treatment; quantification was checked for each sample using probes for GAPDH mRNA. The oligonucleotides used for IκBα are described in “Materials and methods.” The values represent mean ± SEM. (B) To investigate the time kinetics of LPS-induced expression of IκBα, mRNA cells were treated up to 180 minutes with 1 μg/mL LPS (n = 3). A representative figure is shown.
Relevant up-regulation of IκBα mRNA by LPS and expression of oscillatory kinetics under prolonged stimulation. (A) PMNs were incubated with 1 μg/mL LPS, 20 ng/mL GM-CSF (CSF), or 10–7 M DEX for the indicated time points and synthesis of IκBα mRNA was assessed by quantitative RT-PCR (n = 6 for LPS; n = 3 for GM-CSF and DEX). Total RNAs were isolated according to a Qiagen protocol including DNase treatment; quantification was checked for each sample using probes for GAPDH mRNA. The oligonucleotides used for IκBα are described in “Materials and methods.” The values represent mean ± SEM. (B) To investigate the time kinetics of LPS-induced expression of IκBα, mRNA cells were treated up to 180 minutes with 1 μg/mL LPS (n = 3). A representative figure is shown.
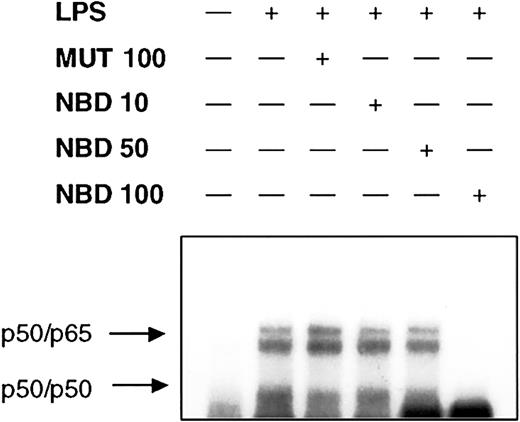
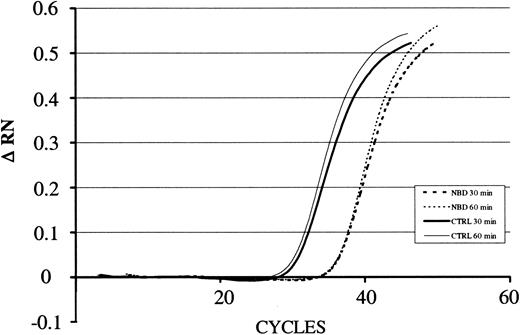
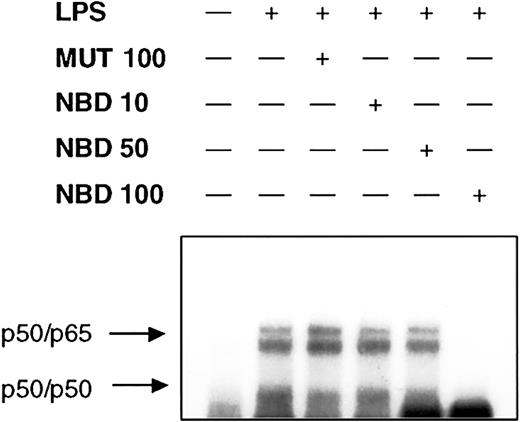
We next investigated whether or not delivery of the NBD peptide by the TAT-PTD would block LPS-induced NF-κB activation in PMNs. Cells were preincubated for 60 minutes with increasing concentrations of TAT-NBD or mutant control prior to stimulation with LPS. Cells were harvested 90 minutes after addition of LPS and EMSA were performed on nuclear extracts. TAT-NBD, but not the control peptide, inhibited NF-κB activation (Figure 6). Furthermore, we observed that 50 and 100 μM TAT-NBD prevented LPS-induced IκBα degradation using Western blotting (n = 2, data not shown). To study whether or not NBD also inhibited NF-κB–dependent gene activation, we performed 3 experiments exploring the effect on IκBα mRNA expression. By RT-PCR, we observed that TAT-NBD, but not the control peptide, prevented LPS-induced IκBα up-regulation (Figure 7).
NEMO-binding domain (NBD) peptide abrogates LPS-induced NF-κB mobilization. Cells (2 × 106) were either left untreated or were incubated with control peptide (MUT, 100 μM) or increasing concentrations of NBD (10, 50, and 100 μM) for 60 minutes before treatment with 1 μg/mL LPS for 90 minutes. NF-κB binding activity was assessed by EMSA. Pretreatment with 100 μM TAT-NBD led to the abrogation of LPS-induced NF-κB activity. A representative figure of 3 independent experiments is depicted.
NEMO-binding domain (NBD) peptide abrogates LPS-induced NF-κB mobilization. Cells (2 × 106) were either left untreated or were incubated with control peptide (MUT, 100 μM) or increasing concentrations of NBD (10, 50, and 100 μM) for 60 minutes before treatment with 1 μg/mL LPS for 90 minutes. NF-κB binding activity was assessed by EMSA. Pretreatment with 100 μM TAT-NBD led to the abrogation of LPS-induced NF-κB activity. A representative figure of 3 independent experiments is depicted.
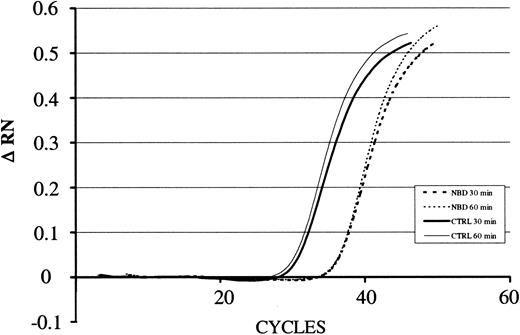
LPS-induced IκBα mRNA expression in the presence of NBD or control peptide. For quantification of the amount of RNA present in the various samples, the fluorescence signal was measured at each PCR cycle and the increase in the fluorescence normalized reporter signal (RN) documented in an amplification plot. The increase in the fluorescence normalized reporter signal is depicted on the y-axis (ΔRN). TAT-NBD (100 μM) but not the control (CTRL) peptide inhibited LPS-induced (1 μg/mL) up-regulation of IκBα. A typical study of 3 independent experiments is given.
LPS-induced IκBα mRNA expression in the presence of NBD or control peptide. For quantification of the amount of RNA present in the various samples, the fluorescence signal was measured at each PCR cycle and the increase in the fluorescence normalized reporter signal (RN) documented in an amplification plot. The increase in the fluorescence normalized reporter signal is depicted on the y-axis (ΔRN). TAT-NBD (100 μM) but not the control (CTRL) peptide inhibited LPS-induced (1 μg/mL) up-regulation of IκBα. A typical study of 3 independent experiments is given.
LPS is not the only inflammatory mediator during inflammation. In fact, LPS itself stimulates other proinflammatory cytokines such as TNF-α resulting in further increase of NF-κB activity. We performed additional experiments exploring the effect of TAT-NBD on TNF-α–induced NF-κB activation. By EMSA we found that 100 μM TAT-NBD inhibited NF-κB activity in cells stimulated with 20 ng/mL TNF-α for 20 minutes (n = 3, data not shown).
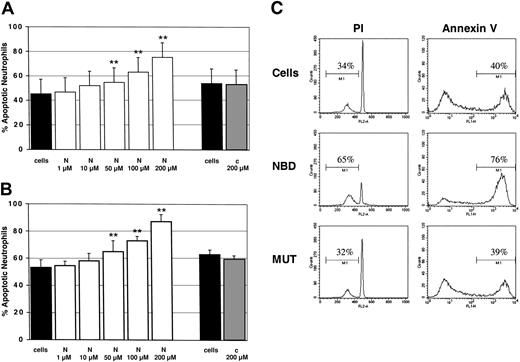
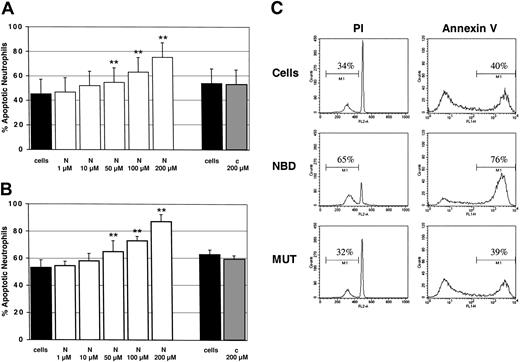
Finally, we investigated whether or not the TAT-NBD construct would manipulate PMN function. To evaluate the effect on constitutive apoptosis, PMNs were incubated for 20 hours with increasing TAT-NBD concentrations ranging from 1 to 200 μM, or with the TAT-control peptide at the highest (200 μM) concentration. Flow cytometry indicated a dose-dependent increase in the percentage of apoptotic PMNs by specific inhibition of NF-κB with TAT-NBD (Figure 8). This effect was observed by 2 independent methods assaying either DNA fragmentation (propidium iodide staining, Figure 8A) or cell surface events (annexin V staining, Figure 8B). A significant increase of apoptosis occurred with 50, 100, and 200 μM of TAT-NBD, respectively. A typical experiment, where both assays were performed in parallel, is shown in Figure 8C. In contrast, no effect was observed when the mutant control peptide was used. These data demonstrate that TAT-NBD accelerates constitutive PMN apoptosis in a dose-dependent fashion. This effect is somewhat unexpected given the fact that unstimulated PMNs did not show any detectable basal NF-κB activity. Toxicity of TAT-NBD was excluded because cell viability was found to be more than 95% by trypan blue exclusion.
Constitutive PMN apoptosis in the presence of NBD or control peptide. Cells were incubated with buffer control (cells), TAT-NBD (N), or mutant control peptide (c). After 20 hours, cells were harvested and the percentage of apoptotic PMNs was assayed by flow cytometry using (A) propidium iodide (PI; n = 4) or (B) annexin V staining (n = 4). **P < .01 when compared with untreated cells. The values represent mean ± SEM. (C) A typical experiment with both assays performed in parallel is depicted using a peptide concentration at 100 μM. The data indicate that TAT-NBD (NBD) accelerated apoptosis in a dose-dependent fashion over a range of 1 to 200 μM. In contrast, no effect was seen in the presence of a mutant control peptide (MUT). The percentage of apoptotic cells is indicated by the marker.
Constitutive PMN apoptosis in the presence of NBD or control peptide. Cells were incubated with buffer control (cells), TAT-NBD (N), or mutant control peptide (c). After 20 hours, cells were harvested and the percentage of apoptotic PMNs was assayed by flow cytometry using (A) propidium iodide (PI; n = 4) or (B) annexin V staining (n = 4). **P < .01 when compared with untreated cells. The values represent mean ± SEM. (C) A typical experiment with both assays performed in parallel is depicted using a peptide concentration at 100 μM. The data indicate that TAT-NBD (NBD) accelerated apoptosis in a dose-dependent fashion over a range of 1 to 200 μM. In contrast, no effect was seen in the presence of a mutant control peptide (MUT). The percentage of apoptotic cells is indicated by the marker.
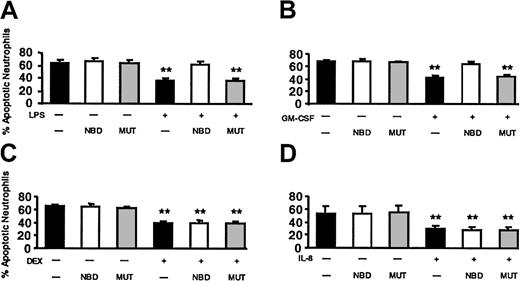
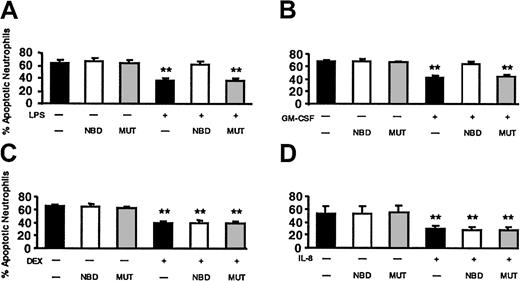
Next, we tested the effect of NBD on LPS-delayed apoptosis. Based on the dose-response curve (Figure 8) and additional dose-finding experiments (data not shown), we selected 10 μM NBD peptide. This concentration had no effect on constitutive apoptosis and completely abrogated the effect of LPS on PMN apoptosis (Figure 9). No effect was observed when the mutant control peptide was used. TAT-NBD did not prevent DEX-induced delay of apoptosis, but abrogated delayed apoptosis by GM-CSF (n = 5 for each compound). The latter result was surprising and prompted us to test the effect of TAT-NBD on an additional neutrophil apoptosis-delaying cytokine, namely IL-8. IL-8 does not activate NF-κB, and we showed previously that IL-8 leads to a delay of apoptosis in PMNs using ERK- and phosphatidylinositol-3-kinase (PI3K)–mediated signal transduction.26-28 The results indicate that TAT-NBD had no effect on delayed apoptosis by IL-8 (n = 4). The results obtained with DEX and IL-8 exclude an unspecific effect of the TAT-NBD. All annexin V data shown in Figure 9 were confirmed by the propidium iodide method (data not shown). Furthermore, the effect of NF-κB inhibition on TNF-α–delayed apoptosis was investigated. TNF-α (20 ng/mL) inhibited apoptosis after 20 hours (68.2% ± 4.7% apoptotic cells in untreated samples versus 47.0% ± 3.6% with TNF-α) and this effect was abrogated by 10 μM TAT-NBD (71.4% ± 2.7%, n = 2).
The effect of NBD on LPS-, GM-CSF–, DEX-, and IL-8–delayed apoptosis. Cells were incubated with buffer control (▪), 10 μM TAT-NBD (□), or the same concentration of a mutant control peptide (▦) in the absence or presence of 100 ng/mL LPS (A), 20 ng/mL GM-CSF (B), 10–7 M DEX (C), or 100 nM IL-8 (D). After 20 hours cells were harvested and the percentage of apoptotic PMNs was assayed by flow cytometry using annexin V staining (n = 5 for LPS, GM-CSF, DEX; n = 4 for IL-8). The data indicate that LPS did not inhibit apoptosis in the presence of TAT-NBD, whereas significant delay of apoptosis occurred in the presence of buffer control or the mutant control peptide. Although TAT-NBD had no effect on the delayed apoptosis by DEX and IL-8, TAT-NBD prevented delayed apoptosis by GM-CSF (**P < .01 when compared to untreated cells).
The effect of NBD on LPS-, GM-CSF–, DEX-, and IL-8–delayed apoptosis. Cells were incubated with buffer control (▪), 10 μM TAT-NBD (□), or the same concentration of a mutant control peptide (▦) in the absence or presence of 100 ng/mL LPS (A), 20 ng/mL GM-CSF (B), 10–7 M DEX (C), or 100 nM IL-8 (D). After 20 hours cells were harvested and the percentage of apoptotic PMNs was assayed by flow cytometry using annexin V staining (n = 5 for LPS, GM-CSF, DEX; n = 4 for IL-8). The data indicate that LPS did not inhibit apoptosis in the presence of TAT-NBD, whereas significant delay of apoptosis occurred in the presence of buffer control or the mutant control peptide. Although TAT-NBD had no effect on the delayed apoptosis by DEX and IL-8, TAT-NBD prevented delayed apoptosis by GM-CSF (**P < .01 when compared to untreated cells).
Our data indicate that 10 μM TAT-NBD completely prevented LPS-delayed apoptosis, whereas no effect of this concentration was detectable in the gel shift assays. We considered the longer incubation time with the peptide during the apoptosis assay as a possible explanation for this finding. Thus, we prolonged the LPS incubation to a maximum of 6 hours and observed by EMSA that 10 μM TAT-NBD, but not the control peptide, clearly diminished LPS-induced NF-κB activity after 4 and 6 hours (n = 2, data not shown).
The increase of constitutive apoptosis with the TAT-NBD raises the suggestion of a basal NF-κB activity that was not detected by EMSA. We therefore established the IKK activity assay and observed the presence of a basal activity in resting cells. LPS (Figure 10) but not GM-CSF (data not shown) resulted in an increase of this basal IKK activity. We then assessed the effect of increasing concentrations of TAT-NBD (10-100 μM) on both basal and LPS-induced IKK activity. Whereas 100 μM TAT-Ctrl (MUT) had no effect, TAT-NBD decreased constitutive and LPS-induced activity in a dose-dependent manner.
TAT-NBD inhibited basal and LPS-induced IKK activity in a dose-dependent manner. The effect of TAT-NBD on constitutive and LPS-induced IKK activity was studied with in vitro kinase assay. Cells (6 × 106) pretreated with either buffer control, TAT-NBD (NBD), or the mutant control (MUT) were stimulated with 1 μg/mL LPS or left untreated. Whole cell lysates were immunoprecipitated with a monoclonal IKKα antibody. Precipitates were incubated with 1 μg GST-IκBα and 3 μCi (0.111 MBq) [γ-32]ATP. Samples were boiled and subjected to 12% SDS-PAGE and autoradiography. Equal loading was confirmed by IKKα Western blot.
TAT-NBD inhibited basal and LPS-induced IKK activity in a dose-dependent manner. The effect of TAT-NBD on constitutive and LPS-induced IKK activity was studied with in vitro kinase assay. Cells (6 × 106) pretreated with either buffer control, TAT-NBD (NBD), or the mutant control (MUT) were stimulated with 1 μg/mL LPS or left untreated. Whole cell lysates were immunoprecipitated with a monoclonal IKKα antibody. Precipitates were incubated with 1 μg GST-IκBα and 3 μCi (0.111 MBq) [γ-32]ATP. Samples were boiled and subjected to 12% SDS-PAGE and autoradiography. Equal loading was confirmed by IKKα Western blot.
These data provide firm evidence that constitutive and LPS-delayed apoptosis are controlled by NF-κB and that the TAT-PTD can be used to specifically target intracellular components of this pathway. Furthermore, this strategy allows manipulation of PMN apoptosis.
Discussion
We demonstrate the possibility of transducing a specific NF-κB inhibitory peptide synthesized to a PTD of the HIV-TAT protein into human PMNs. Our results indicate that this peptide entered virtually all cells and completely blocked LPS-induced NF-κB activity. Furthermore, we provide firm proof that this strategy presents a feasible approach to specifically target intracellular key proteins and ultimately to control biologic functions, as demonstrated for NF-κB–dependent apoptosis, even in human PMNs. This tool presents an approach superior to any other method reported in PMNs, such as electroporation, lipofectin, or hypotonic shock. We are aware that our in vitro results are necessarily artificial and caution needs to be warranted to translate these observations to in vivo situations.
We selected NF-κB–dependent signaling because of the powerful regulatory role in PMN apoptosis. Resolution of inflammation depends largely on neutrophil apoptosis, and the removal of the apoptotic cells in a noninflammatory manner.29 We had to overcome several problems that complicate studies of NF-κB DNA-binding activity in PMNs. For example, the high degree of proteolytic activity led to rapid NF-κB degradation during nuclear extract preparation and undesired protein degradation still occurred with different protocols. A modified protocol that included the detergent NP40 still led to nuclear degradation, even when only small concentrations were added. McDonald and coworkers already performed extensive optimizing; they recommended modified nuclear extract preparation, including nitrogen cavitation.21 Disruption of the cells was sufficient with nitrogen cavitation; however, a high cell number (30 × 106) and a large assay volume were necessary. We finally established a protocol using hypotonic/hypertonic buffers, omitting NP40, and avoided high centrifugation steps after cell membrane disruption to prevent pelleting granules containing proteolytic enzymes. With these adjustments, we were able to investigate NF-κB activity by EMSA and demonstrated the release of a p50/p50 homodimer and a p50/p65 heterodimer in our supershift assays.
Our data, obtained from parallel studies, demonstrate that constitutive PMN apoptosis is delayed by LPS, GM-CSF, and DEX. These results are in agreement with previous reports.28,30 Recently, Sabroe et al provided data showing that the inhibitory effect of LPS on PMNs in vitro is amplified by very small numbers of contaminating monocytes.31 Moreover, we found that only LPS, but not GM-CSF or DEX, caused the cytoplasmic inhibitor of NF-κB, IκBα, to be degraded, thereby activating NF-κB nuclear transmigration and inducing NF-κB–dependent gene transcription, as demonstrated for IκBα mRNA expression. Time-course studies of LPS-induced IκBα mRNA expression revealed an oscillatory kinetic. Hoffmann and colleagues recently developed a computational model that points out the control of NF-κB activation by the coordinated degradation and synthesis of IκB proteins.32 The fact that IκBα mediates rapid NF-κB activation and a strong negative feedback regulation resulted in an oscillatory NF-κB activation profile. Thus, it is conceivable that these kinetics lead to an alternating up- and down-regulation of IκBα mRNA, providing a possible explanation for the results observed in our study. Another possibility could be that activation of NF-κB leads to the up-regulation of other transcription repressors as a negative feedback mechanism or that the composition at the IκBα promoter site changes during prolonged activation, leading to an unstable expression. Only little is known about mRNA stability. Recently, it was shown that UV light stabilizes IκBα mRNA expression in murine fibroblasts.33
Apparently, GM-CSF and DEX delay apoptosis by other pathways than NF-κB. For example, GM-CSF may delay apoptosis by STAT-dependent regulation of the antiapoptotic Mcl-1 protein and also by up-regulation of A1 mRNA expression, an antiapoptotic member of the BCL2 family.34 We and others demonstrated the importance of the PI3K pathway in GM-CSF–mediated delay of apoptosis in human neutrophils.28,35 DEX also delayed apoptosis in our experiments using human PMNs. This observation is in contrast to studies in T cells and in myeloid lineages, where DEX rapidly induced apoptosis.36 Recently, a molecular explanation for this observation was reported.37 Strickland et al37 characterized 2 isoforms of the human glucocorticoid receptor, namely GRα and GRβ. They showed that DEX-induced apoptosis requires the GRα and that is less expressed in human granulocytes than in peripheral blood mononuclear cells or mouse PMNs. Human PMNs revealed an inversion of the GRα/GRβ ratio, where GRβ protects from apoptotic processes. The requirement of transcription activity and protein synthesis for DEX-delayed apoptosis in human PMNs was indicated by Cox et al.38 Our results exclude NF-κB as a central transcription factor mediating delayed apoptosis by DEX. Further studies are needed to identify transcription factors that control the effect of DEX on PMN apoptosis.
To specifically block NF-κB–dependent signaling, we selected the NEMO-binding domain (NBD) peptide that was first described by May et al.20 These authors showed that NBD specifically blocks the interaction of IKKγ with the IKK complex. No effect on other transcription factors, like Oct-1 for example, was observed. We did not use the bacterial expression system reported by May and coworkers, but instead generated a linear peptide consisting of the NBD and an NH2-terminal TAT-PTD sequence. The transduction method and the specific inhibitory peptide used in our study extend the results from previous neutrophil studies using less specific NF-κB inhibitors.16,39,40 Our results indicate that intracellular delivery of TAT-NBD completely blocked LPS-induced IKK kinase activity, NF-κB–binding activity, and NF-κB–dependent gene transcription. Inhibition of NF-κB by TAT-NBD resulted in accelerated constitutive PMN apoptosis. This effect could be explained by the existence of low-level basal NF-κB activity as shown by IKK activity assays. Furthermore, possibly TAT-NBD inhibits constitutive active Rel-dimers of NF-κB that cannot be detected in the shift assay, suggesting the presence of unknown Rel family members in PMNs with permanent NF-κB activity. Furthermore, TAT-NBD abrogated LPS-delayed apoptosis. TAT-NBD did not prevent DEX- and IL-8–mediated delay of apoptosis, but abrogated delayed apoptosis by GM-CSF. The data obtained with DEX and IL-8 exclude an unspecific effect of the TAT-NBD. Nevertheless, we currently cannot explain the unexpected effect of TAT-NBD on delayed apoptosis by GM-CSF. We provided several lines of evidence that GM-CSF does not increase IKK or NF-κB activity. It is still conceivable that GM-CSF needs basal IKK/NF-κB activity to up-regulate antiapoptic genes, but does not require events that are further downstream in this pathway. In fact, Li and colleagues obtained DNA microarray data showing that LPS induces IKK-dependent target genes in the 70Z/3 murine pre-B cell line but not in the IKKγ-deficient variant of 1.3E2 cells. The authors described genes that were directly dependent on IKKγ (NEMO). These genes were not affected in LPS-treated 70Z/3 cells after transfection with an IκBα superrepressor.41 Thus, NEMO may regulate a class of genes independent of classical NF-κB.41 (D. Krappmann, unpublished data, November 2002). The role of the IKK complex in GM-CSF signaling is intriguing and will be further investigated in future studies. In any event, our data now provide firm proof of the functional significance of NF-κB in PMN apoptosis and open new avenues to investigate signaling in PMNs.
Prepublished online as Blood First Edition Paper, May 22, 2003; DOI 10.1182/blood-2002-09-2960.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Jana Czychi for excellent technical assistance.

![Figure 10. TAT-NBD inhibited basal and LPS-induced IKK activity in a dose-dependent manner. The effect of TAT-NBD on constitutive and LPS-induced IKK activity was studied with in vitro kinase assay. Cells (6 × 106) pretreated with either buffer control, TAT-NBD (NBD), or the mutant control (MUT) were stimulated with 1 μg/mL LPS or left untreated. Whole cell lysates were immunoprecipitated with a monoclonal IKKα antibody. Precipitates were incubated with 1 μg GST-IκBα and 3 μCi (0.111 MBq) [γ-32]ATP. Samples were boiled and subjected to 12% SDS-PAGE and autoradiography. Equal loading was confirmed by IKKα Western blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/6/10.1182_blood-2002-09-2960/6/m_h81834919010.jpeg?Expires=1769219778&Signature=KZXT6GoTgefHWwNypPDUZDhIc3XCOaclcxkJQq674-EZFF8jOE8MkD~-uRNnR1s23IW9AILMUfg6PFjuLkQ3sCXjYzz4XqIDDiNuhG1gK~TU37hpusaBrGtTpzC~yVIb0PE7apjPA3dNmNAZUBzPIyIdldC9QRrpxR9bc3CT5ySCq-y~joQqZxgu932oYQBdnyHaxb-2LEa0es-cZMrLM2P9HPRZHnNtfSwxtcLGPpvo1LfautHRVvNYW2BdqhmKo1VBo5h~-PbieBViNv~xypqpLEvFdzSJ2tslxSDMIt3VnD-SJBjsGjNI604E57S5jpyz3Mn0VdVTOxD4gql-VA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 10. TAT-NBD inhibited basal and LPS-induced IKK activity in a dose-dependent manner. The effect of TAT-NBD on constitutive and LPS-induced IKK activity was studied with in vitro kinase assay. Cells (6 × 106) pretreated with either buffer control, TAT-NBD (NBD), or the mutant control (MUT) were stimulated with 1 μg/mL LPS or left untreated. Whole cell lysates were immunoprecipitated with a monoclonal IKKα antibody. Precipitates were incubated with 1 μg GST-IκBα and 3 μCi (0.111 MBq) [γ-32]ATP. Samples were boiled and subjected to 12% SDS-PAGE and autoradiography. Equal loading was confirmed by IKKα Western blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/6/10.1182_blood-2002-09-2960/6/m_h81834919010.jpeg?Expires=1769219779&Signature=Rx58UDIGiUm7NXL6NaJvdr5MTPPfrJB8SWAuypS49dSg1M~8g5H7PKHsQvLV8kfHLbz46qgq6O~aNIpN9Z7vK~4srHJTj7a65OCV0C1U2crlKGrozWAaYM4zfp3a2jjPArlURYjeSUVJBQB59gb0agM0aFZrFsDZ93RFQkb1ovLHWFHuHGzJLXdjSd68CvLaTBFnhek90M9bV2eXs7sJw1bw28HtbWeXxyxB4febewNobM7u4ubUtIm5kX-yxAj4DvxrICy3Zv-dI2xXws~lMc5tEKeJvT1lhOOoT1plVnsS7I1cPTY3x3q53mSU736k9zzJgu~GQQNBGTzzUf58eQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)