Abstract
Decreased expression of c-MPL protein in platelets, increased expression of polycythemia rubra vera 1 (PRV-1) and nuclear factor I-B (NFIB) mRNA in granulocytes, and loss of heterozygosity on chromosome 9p (9pLOH) were described as molecular markers for myeloproliferative disorders (MPDs). To assess whether these markers are clustered in subgroups of MPDs or represent independent phenotypic variations, we simultaneously determined their status in a cohort of MPD patients. Growth of erythropoietin-independent colonies (EECs) was measured for comparison. We observed concordance between EECs and PRV-1 in MPD patients across all diagnostic subclasses, but our results indicate that EECs remain the most reliable auxiliary test for polycythemia vera (PV). In contrast, c-MPL, NFIB, and 9pLOH constitute independent variations. Interestingly, decreased c-MPL and elevated PRV-1 also were observed in patients with hereditary thrombocythemia (HT) who carry a mutation in the thrombopoietin (TPO) gene. Thus, altered c-MPL and PRV-1 expression also can arise through a molecular mechanism different from sporadic MPD.
Introduction
Previous reports suggested that decreased expression of c-MPL protein in platelets and elevated expression of polycythemia rubra vera 1 (PRV-1) mRNA in granulocytes are characteristic features of patients with polycythemia vera (PV).1,2 These findings raised hopes that assessing expression of c-MPL and PRV-1 may replace the need for determining the growth of erythropoietin-independent colonies (EECs), a valuable but technically demanding assay for PV.3-5 Since low c-MPL and high PRV-1 also were detected in some patients with essential thrombocythemia (ET) and chronic idiopathic myelofibrosis (IMF),1,2,6-8 these markers might define subsets within these myeloproliferative disorder (MPD) entities. Recently, 9pLOH was described as the most frequent chromosomal aberration in PV patients, and increased expression of mRNA for the transcription factor nuclear factor I-B (NFIB) was found in some patients with 9pLOH.9 To determine whether these markers constitute independent phenotypic variations or appear clustered in subgroups of MPD patients, we simultaneously determined the c-MPL, PRV-1, EEC, NFIB, and 9pLOH status in a cohort of 44 MPD patients and 18 healthy control individuals.
Study design
The study was approved by the Ethics Committee of Basel. Blood was obtained with written consent. The diagnosis of MPD subtypes was assigned using the World Health Organization (WHO) criteria.10
Protein, RNA, and DNA analyses
All blood samples were processed within 1-4 hours of drawing. Isolation of granulocyte RNA and DNA is described elsewhere.9 RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Platelets were purified using the sepharose gel-filtration method.11 c-MPL protein expression was determined by immunoblot analysis using the polyclonal rabbit antibody (CTP7) specific for the C-terminus of human c-MPL (kindly provided by Dr A. Moliterno and Dr J. Spivak, Johns Hopkins University, Baltimore, MD). To normalize for platelet protein loading, the membranes were reprobed using a monoclonal antibody against human CD61 (BD Biosciences, San Jose, CA). Thrombopoietin (TPO) serum levels were determined using the TPO-Quantikine ELISA (enzyme-linked immunosorbent assay) (R&D Systems, Minneapolis, MN).
Real-time polymerase chain reaction
Total RNA (2 μg) was reverse transcribed after random hexamer priming. To prevent influence from genomic DNA amplification, the primers for RPL19, PRV-1, and NFIB were designed across exon-intron junctions. The primers for RPL19 were GATGCCGGAAAAACACCTTG, TGGCTGTACCCTTCCGCTT, CCTATGCCCATGTGCCTGCCCTT (probe); for PRV-1: CCCCAGCAGACCCAGGA, TTGTCCCCTCCAGACAGCC, CCATAGACAAGCAGACTGGGCACCTCAA (probe); and for NFIB: CAGTCCACAAACCAGCCAGTC, GCCGGTAAGATGGGTGTCCTA, GAAAGGAACCAAGCTAGCCCAGGTACCA (probe). The probes were dual-labeled with 5′-6-carboxyfluorescein (FAM) and 3′-carboxytetramethylrhodamine (TAMRA). The primers for MPL used for SYBR-PCR were AGCCCTGAGCCCGCC and TCCACTTCTTCACAGGTATCTGAGA. The ΔCT values were derived by subtracting the threshold cycle (CT) values for PRV-1, NFIB, and c-MPL from the CT value for ribosomal protein L19 (RPL19), which served as an internal control.12 All reactions were run in duplicate using the ABI 7000 Sequence Detection System (Applied Biosystems, Foster City, CA).
EEC assay
The clonogenic cultures for erythropoietin-independent colony formation (EEC assay) were performed using commercial reagents (Stem Cell Laboratories, Vancouver, BC, Canada).
Detection of 9pLOH
Three highly polymorphic microsatellite markers D9S1779, D9S157, and D9S161 were used to detect LOH in granulocyte DNA samples as described.9
Results and discussion
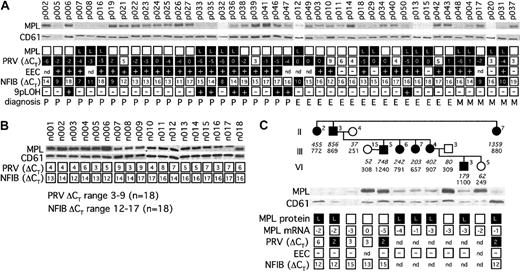
To determine whether alterations in platelet c-MPL protein expression, granulocyte PRV-1, and NFIB mRNA levels, growth of EECs, and the presence of 9pLOH represent independent phenotypic variations, we examined 44 patients with MPD (23 PV, 15 ET, and 6 IMF) (Figure 1A). As controls, 18 healthy individuals were included (Figure 1B). Decreased expression of c-MPL protein was found in 30% of patients with PV (7 of 23), 40% of ET (6 of 15) and 67% of IMF (4 of 6). Thus, c-MPL cannot be used as a diagnostic test for PV. To assess whether lower expression of c-MPL is specific for MPD, we examined a family with hereditary thrombocythemia (HT). In this family, thrombocytosis is caused by elevated TPO serum levels due to a splice donor mutation in the TPO gene.13 We found lower expression of c-MPL protein in 7 of 8 affected individuals (88%), despite normal c-MPL mRNA levels (Figure 1C). Hence, decrease of c-MPL protein also can occur in patients who display sustained thrombocytosis caused by a molecular mechanism different from sporadic MPD.
Summary of MPD marker analysis. (A) Cohort of 44 MPD patients. Open squares signify normal findings, black squares indicate abnormal values. Expression of c-MPL protein from platelets was detected by Western blot analysis and compared with CD61 to normalize for platelet protein loading. Unique patient numbers were placed above the corresponding lanes. L indicates decreased expression of c-MPL protein. PRV-1 and NFIB mRNA levels were determined by real-time PCR. Numbers indicate the ΔCT values (see “Study design”). Note that high numeric ΔCT values indicate low or normal abundance of mRNA (open squares), whereas low or negative ΔCT values signify elevated levels of expression (black squares). Presence (+) or absence (–) of EEC or 9pLOH is indicated. nd indicates not determined; P, polycythemia vera; E, essential thrombocythemia; and M, chronic idiopathic myelofibrosis. (B) Healthy controls. (C) Hereditary thrombocythemia. Individuals within the pedigree are placed above the corresponding lanes and are numbered as by Wiestner et al.13 Black symbols indicate affected individuals. Serum TPO concentration in pg/mL (numbers in italic) and platelet count × 109/L (numbers in regular style) are shown below each individual.
Summary of MPD marker analysis. (A) Cohort of 44 MPD patients. Open squares signify normal findings, black squares indicate abnormal values. Expression of c-MPL protein from platelets was detected by Western blot analysis and compared with CD61 to normalize for platelet protein loading. Unique patient numbers were placed above the corresponding lanes. L indicates decreased expression of c-MPL protein. PRV-1 and NFIB mRNA levels were determined by real-time PCR. Numbers indicate the ΔCT values (see “Study design”). Note that high numeric ΔCT values indicate low or normal abundance of mRNA (open squares), whereas low or negative ΔCT values signify elevated levels of expression (black squares). Presence (+) or absence (–) of EEC or 9pLOH is indicated. nd indicates not determined; P, polycythemia vera; E, essential thrombocythemia; and M, chronic idiopathic myelofibrosis. (B) Healthy controls. (C) Hereditary thrombocythemia. Individuals within the pedigree are placed above the corresponding lanes and are numbered as by Wiestner et al.13 Black symbols indicate affected individuals. Serum TPO concentration in pg/mL (numbers in italic) and platelet count × 109/L (numbers in regular style) are shown below each individual.
PRV-1 mRNA was elevated in 91% of patients with PV (21 of 23), 67% of ET (10 of 15), and 67% of IMF (4 of 6), whereas growth of EEC was present in 100% of patients with PV (20 of 20), 69% of patients with ET (9 of 13), and 60% of patients with IMF (3 of 5). Our results show a higher detection rate of PV than a recently published study, which reported that only 69% of PV patients (7 of 13) had elevated PRV-1.14 This discrepancy might be due to the smaller number of patients examined in the previous report. Using the Agilent 2100 Bioanalyzer, we excluded degradation of the RNA as a potential explanation for low PRV-1 in EEC-positive individuals. Another difference between the 2 studies is that our PRV-1 assay is based on primers that span exon-intron boundaries and should therefore be less influenced by DNA contamination in the patient RNA samples. Irrespective of the MPD subtype, we observed a strong correlation between PRV-1 and EEC: 84% of patients with EECs had elevated PRV-1 (27 of 32) and, conversely, 94% of patients with elevated PRV-1 were EEC positive (29 of 31). Interestingly, we also detected increased expression of PRV-1 in 3 of 4 affected HT family members who were available for analysis (Figure 1C). PRV-1 expression in these individuals did not reach the very high levels observed in most patients with sporadic MPD. Cytokines can influence PRV-1 expression, for example, treatment of normal granulocytes with granulocyte colony-stimulating factor increased PRV-1 mRNA.2 It remains to be examined whether elevated TPO levels in HT patients might indirectly influence PRV-1. Importantly, EECs were negative in all affected family members (Figure 1C). Thus, EECs remain the most reliable auxiliary diagnostic assay for PV.
Loss of heterozygosity on the short arm of chromosome 9 (9pLOH) was recently reported to be the most frequent genetic lesion in PV, affecting approximately 30% of PV patients.9 In comparison, deletion of 20q and various chromosome 9 aberrations were previously reported in 15% and 21% of PV patients, respectively.15,16 In our present study, 7 of 23 PV patients (30%) displayed 9pLOH, confirming the previously observed 9pLOH frequency. The 9pLOH also was present in 1 patient with ET. Interestingly, all 8 individuals with 9pLOH were EEC positive and also displayed elevated PRV-1. Expression of NFIB was hypothesized to be a consequence of 9pLOH.9 Our study detected increased NFIB expression in 8 MPD patients with EEC (5 PV, 1 ET, and 2 IMF), but none of these individuals had 9pLOH, indicating that increased expression of the NFIB gene is independent of the presence of 9pLOH. The correlation between 9pLOH and increased NFIB in the original report was most likely a coincidence within a small cohort of 3 patients. No increase in NFIB mRNA was noted in HT family members (Figure 1C).
The concordance between EEC and PRV-1 markers in MPD patients across all diagnostic subclasses suggests that they might be caused by a common molecular mechanism. In contrast, decreased expression of c-MPL appears to be an independent phenotypic variant. Interestingly, low c-MPL and high PRV-1 also can occur secondary to elevated TPO in patients with HT. Increased NFIB expression and presence of 9pLOH are independent of each other but were detected only in patients with EEC and/or high PRV-1. Larger cohort studies will be necessary to determine whether MPD subgroups defined by these molecular markers differ in respect to clinical outcome, for example, leukemic transformation or hemorrhagic and thrombotic complications.
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2003-03-0744.
Supported by grants from the Swiss National Science Foundation, the Swiss Cancer League, and the Lichtenstein Stiftung (R.K.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Patricia Frank and Michael Kirsch for technical assistance, and Alois Gratwohl and Aleksandra Wodnar-Filipowicz for helpful comments on the manuscript.