Abstract
Congenital afibrinogenemia is a rare autosomal recessive disorder characterized by complete absence of detectable fibrinogen. We previously identified the first causative mutations for this disease: a homozygous deletion of approximately 11 kb of the fibrinogen α-chain gene (FGA). Subsequent studies revealed that the great majority of afibrinogenemia mutations are localized in FGA, but mutations were also found in FGG and FGB. Apart from 3 missense mutations identified in the C-terminal portion of FGB, all fibrinogen gene mutations responsible for afibrinogenemia are null. In this study, a young boy with afibrinogenemia was found to be a compound heterozygote for 2 mutations in FGB: an N-terminal nonsense mutation W47X (exon 2) and a missense mutation (G444S, exon 8). Coexpression of the FGB G444S mutant cDNA in combination with wild-type FGA and FGG cDNAs demonstrated that fibrinogen molecules containing the mutant β chain are able to assemble but are not secreted into the media, confirming the pathogenic nature of the identified mutation. (Blood. 2003;102:4413-4415)
Introduction
Congenital afibrinogenemia (Mendelian Inheritance in Man, MIM no. 202400) is a rare autosomal recessive disorder characterized by complete absence of detectable fibrinogen in circulation. Uncontrolled bleeding after birth from the umbilical cord is the most frequent symptom; joint, mucosal and intracerebral bleeding, splenic rupture, or miscarriage can occur throughout life.1 Since the original description of the disorder in 1920,2 more than 150 cases have been reported.
Fibrinogen is a 340-kDa glycoprotein predominantly synthesized by hepatocytes; it is composed of 2 sets of 3 homologous polypeptide chains known as Aα, Bβ, and γ chains that assemble to form an hexameric structure (AαBβγ)2. Each polypeptide is encoded by a distinct gene, FGA, FGB, and FGG, respectively, clustered in a region of 50 kb on chromosome 4q28-31.3 Since our identification of the disease locus and of the first described causative mutation, a deletion of approximately 11 kb in FGA gene,4 numerous other mutations have been described in FGA (the great majority of cases), FGG, and FGB.5 Apart from 3 missense mutations in FGB,6,7 all mutations described so far are null (frameshift, nonsense, or splice-site mutations) predicted to cause total lack of the corresponding fibrinogen chain. These missense mutations all lie in the C-terminal portion of the fibrinogen β chain, which is highly conserved among vertebrates, from lamprey to human. For 2 of these, L383R (numbering from the initiator methionine, according to Human Genome Organization [HUGO] recommendations8 ) in exon 7 and G430D in exon 8 (formerly termed L353R and G400D, respectively, numbering after signal peptide cleavage), expression in transfected COS-1 cells followed by quantification of intracellular or secreted fibrinogen products after pulse-chase was performed to prove their causative nature. The mutations led to deficient secretion of the fibrinogen hexameric complex, probably because of impaired Bβ side-chain packing leading to incorrect folding of the mutated chains.6
In addition, 2 nonsense mutations in the same highly conserved region were recently characterized. The first, “fibrinogen Mount Eden”9 or W470X (formerly Trp440Stop) was identified in heterozygosity in an asymptomatic patient following laboratory investigations prior to a liver biopsy for hepatitis C. The second nonsense mutation W467X, 3 codons upstream, was identified in our laboratory in homozygosity in 2 Palestinian sisters with afibrinogenemia.10
In this study, a young patient with afibrinogenemia was found to be a compound heterozygote for 2 mutations in FGB: R47X (exon 2), which has been previously found in an afibrinogenemic patient from Iran,11 and G444S, a missense mutation located in exon 8. The localization of this missense mutation prompted us to perform cotransfection experiments in COS-7 cells as previously described for the W467X mutation10 to assay its consequence on fibrinogen assembly or secretion.
Study design
Description of family
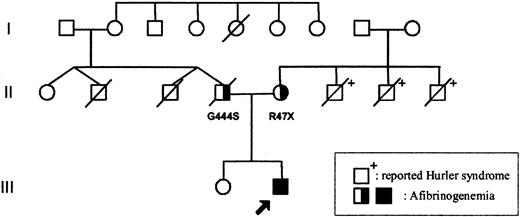
The parents of the patient were unrelated (Figure 1). The mother's family originates from England and the father's family from Germany. Both parents displayed normal prothrombin time (PT) and activated partial thromboplastin time (APTT) values, but their fibrinogen levels were reduced: 1.48 (father) and 2.0 mg/mL (mother, 2 weeks after delivery). Fibrinogen values in family members are shown in Table 1.
Family tree. The arrow indicates the proband. Apart from the father (II.4) who died from a presumed myocardial infarction, no history of abnormal bleeding or thrombosis was reported on either side of the family (individual II.2 died in an accident at age 34; individual II.3 died close to birth). The mother's 3 brothers (II.6, II.7, and II.8) died from reported Hurler syndrome. □ = male; ○, female; and /, deceased.
Family tree. The arrow indicates the proband. Apart from the father (II.4) who died from a presumed myocardial infarction, no history of abnormal bleeding or thrombosis was reported on either side of the family (individual II.2 died in an accident at age 34; individual II.3 died close to birth). The mother's 3 brothers (II.6, II.7, and II.8) died from reported Hurler syndrome. □ = male; ○, female; and /, deceased.
The couple had 2 children; the eldest, a daughter, was asymptomatic for bleeding events and had a normal fibrinogen level. The second child, a son, was born healthy at term, weighing 3 kg after a normal pregnancy. He received routine vitamin K prophylaxis. At 2 days of age he developed bilateral cephalohematomas and became pale and lethargic. His blood was found to be incoagulable with both the PT and APTT being more than 200 seconds and the fibrinogen unmeasurable. He was anemic with a hemoglobin level of 8.4 g/dL. After treatment with fresh-frozen plasma (FFP) and blood, his fibrinogen level was 0.5 mg/mL with complete correction of the PT and APTT, suggesting a diagnosis of severe hypofibrinogenemia or afibrinogenemia. Other coagulation factors were normal on the pretreatment blood sample, and this was confirmed at a later date when treatment was withdrawn for a period. Cryoprecipitate infusions were repeated every 2 to 3 days for 2 weeks to keep the fibrinogen level above 1 mg/mL; subsequently he was treated with regular infusions of a fibrinogen concentrate. A central line was required for his continued treatment, and this was complicated by complete but asymptomatic thrombosis of the upper venous system suspected after 6 months when difficulty was experienced replacing his line by a subcutaneous port. Subsequent magnetic resonance angiography confirmed this. Thrombophilia screening demonstrated normal values for proteins C and S, and antithrombin, and he does not have the factor V Leiden or prothrombotic prothrombin gene (G20210A) mutations. When treatment was stopped for a period of 5 months, his baseline coagulation tests confirmed complete absence of detectable fibrinogen. He required treatment on 3 occasions for nonsevere bleeding usually related to trauma. Treatment was recommenced when he started to walk because he was bruising extensively and was felt to be at risk of intracranial hemorrhage. The central line was removed after a further 2 years during which there was no evidence of further thrombotic events, and he is well at the age of 4 years on fortnightly prophylaxis via peripheral veins.
The child's father died suddenly at 34 years of age, when his son was 8 months of age, from a presumed myocardial infarction. Apart from the father, no history of abnormal bleeding or thrombosis was reported on either side of the family. Coincidentally, the patient's mother lost 3 brothers to reported Hurler syndrome (Figure 1).
Mutation screening
Expression and analysis of the mutations in COS-7 cells
Construction of wild-type and mutant expression vectors, transient cotransfections of COS-7 cells, and conditioned media harvesting and Western blot analysis were performed as previously described.10 The oligonucleotides used for site-directed mutagenesis of the G444 codon were: 5′-GCAGCCAATCCAAACAGCAGATACTACTGGGG-3′ and 5′-CCCCAGTAGTATCTGCTGTTTGGATTGGCTGC-3′. Cells were lysed in Ripa buffer: NaCl 150 mM, Tris (tris(hydroxymethyl)aminomethane)-HCl 50 mM pH 8.0, NP-40 1%, sodium dodecyl sulfate (SDS) 0.1%, sodium deoxycholate 0.5%, phenylmethylsulfonyl fluoride (PMSF) 0.5 mM, with protease inhibitors (Complete, Roche, Basel, Switzerland). Samples were incubated on ice for 30 minutes and centrifuged at 13 000 rpm at 4°C for 30 minutes. Laemmli buffer was added to the supernatant, and the samples were boiled at 95°C for 5 minutes before analysis by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Results and discussion
Sequence analysis of the proband's gDNA revealed no mutation in either FGA or FGG but 2 mutations were found in FGB, both in heterozygosity: an early truncating nonsense mutation in exon 2 (R47X, numbered from the initiator ATG) and a missense G444S substitution in exon 8. The compound heterozygosity was confirmed by analyzing gDNA isolated from the parents; the father was a heterozygous carrier of the G444S mutation and the mother of R47X. Both parents had intermediate or normal fibrinogen levels, at 1.48 and 2.0 mg/mL, respectively.
The R47X is predicted to cause the production of a severely truncated β chain, if the protein is produced at all. Alternatively, the mutation might cause a defect in the FGB mRNA, by aberrant splicing (nonsense-associated alternative splicing) or by affecting the stability of the mRNA through nonsense-mediated mRNA decay.14-16 Whatever the mechanism involved, there is no reasonable doubt that this is a null mutation. Indeed, this mutation was recently identified in homozygosity in an Iranian patient, although no functional analysis was performed to determine the molecular mechanism involved.11 Therefore, we focused on the G444S mutation for further analysis.
The FGB G444S cDNA mutant, obtained by site-directed mutagenesis of the wild-type FGB cDNA, was transiently cotransfected in COS-7 cells with wild-type FGA and FGG, and with normal FGB cDNA (mimicking heterozygous state) or alone (homozygous state). Eighteen hours after transfection, individual fibrinogen chains (under reducing conditions) or assembled hexamers (under nonreducing conditions) in cell extracts and conditioned media were detected by Western blot analysis with a polyclonal antifibrinogen antibody. Cotransfection with the 3 normal cDNAs showed expression of all 3 polypeptides, normal assembly inside the cell, and adequate secretion of the hexamer (Figure 2A). When cotransfections were performed with wild-type FGA and FGG cDNAs and with the mutant FGB G444S cDNA, the 3 polypeptides were expressed and were able to assemble intracellularly to form hexamers. When cotransfections were performed with normal FGA, FGG, and equal amounts of wild-type and mutant FGB cDNAs, the 340-kDa fibrinogen hexamer was still secreted into the media. In contrast, coexpression of wild-type FGA and FGG with mutant FGB cDNA alone eliminated fibrinogen secretion.
Transfection studies. (A) Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs. Samples of cell lysates and culture medium were subjected to 10% SDS-PAGE under reducing conditions or 7.5% SDS-PAGE under nonreducing conditions. The blots were incubated with a polyclonal antihuman fibrinogen antibody and cross-reacting bands were revealed by chemiluminescence, as described.10 Fib indicates purified fibrinogen control; —, COS cells transfected with an empty vector. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated. Lane 1: normal Aα, Bβ, and γ; lane 2: normal Aα, normal Bβ plus mutant G444S Bβ, and normal γ; lane 3: normal Aα, mutant G444S Bβ (only), and normal γ. (B) Multiple alignment of the C-terminal region of fibrinogen Bβ chain. Sequences from human (P02675), mouse (XP_130960), rat (P14480), bovine (P02676), chicken (Q02020), Xenopus (Q91589), and lamprey (P02678) were obtained from the Swiss-Prot database (except the mouse sequence, which was obtained from the National Center for Biotechnology Information [NCBI]) and aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw/). Conserved amino acids are highlighted in gray; arrows indicate the positions of previously reported mutations: L383R, G430D, Y447G, W467X, W470X. The open arrow indicates the position of the mutated glycine residue (G444S).
Transfection studies. (A) Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs. Samples of cell lysates and culture medium were subjected to 10% SDS-PAGE under reducing conditions or 7.5% SDS-PAGE under nonreducing conditions. The blots were incubated with a polyclonal antihuman fibrinogen antibody and cross-reacting bands were revealed by chemiluminescence, as described.10 Fib indicates purified fibrinogen control; —, COS cells transfected with an empty vector. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated. Lane 1: normal Aα, Bβ, and γ; lane 2: normal Aα, normal Bβ plus mutant G444S Bβ, and normal γ; lane 3: normal Aα, mutant G444S Bβ (only), and normal γ. (B) Multiple alignment of the C-terminal region of fibrinogen Bβ chain. Sequences from human (P02675), mouse (XP_130960), rat (P14480), bovine (P02676), chicken (Q02020), Xenopus (Q91589), and lamprey (P02678) were obtained from the Swiss-Prot database (except the mouse sequence, which was obtained from the National Center for Biotechnology Information [NCBI]) and aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw/). Conserved amino acids are highlighted in gray; arrows indicate the positions of previously reported mutations: L383R, G430D, Y447G, W467X, W470X. The open arrow indicates the position of the mutated glycine residue (G444S).
The data demonstrate that the FGB G444S mutation allows intracellular hexamer assembly, but inhibits its secretion into the media. Two other missense mutations6 (G430D, L383R, renumbered according to guidelines for human mutation nomenclature8 ) localized in the C-terminal domain of the Bβ polypeptide were previously identified in afibrinogenemic patients and were also shown to impair protein secretion but not assembly. Finally, a heterozygous missense mutation, R285H, in FGB exon 6, was recently discovered in a hypofibrinogenemic individual displaying an intermediate level of plasma fibrinogen (1.4 mg/mL).17 Because heterozygous carriers of an afibrinogenemia allele can also have intermediate fibrinogen concentrations, we suggest that an individual homozygous for R285H would have afibrinogenemia.
In addition to the 4 missense mutations (G444S included) in the FGB gene so far discovered in afibrinogenemic or hypofibrinogenemic patients and all localized in the C-terminal domain of the polypeptide, 2 nonsense mutations, W467X and W470X, were found in the same domain.9,10 The former has also been shown to inhibit fibrinogen secretion and not assembly, and the latter would be expected to act similarly. These combined studies confirm the necessity of an intact C-terminal domain of the Bβ chain for the process of fibrinogen secretion.
Prepublished online as Blood First Edition Paper, July 31, 2003; DOI 10.1182/blood-2003-06-2141.
Supported by Swiss National Science Foundation (SNF) Grant 31-64987.01, by the Telethon Action Suisse Foundation, and by the Ernst and Lucie Schmidheiny Foundation. M.N.-A. is the recipient of an SNF professorship (grant no. 631-66023).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Luciana Palumbo and Christine Hinard for expert technical assistance.

![Figure 2. Transfection studies. (A) Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs. Samples of cell lysates and culture medium were subjected to 10% SDS-PAGE under reducing conditions or 7.5% SDS-PAGE under nonreducing conditions. The blots were incubated with a polyclonal antihuman fibrinogen antibody and cross-reacting bands were revealed by chemiluminescence, as described.10 Fib indicates purified fibrinogen control; —, COS cells transfected with an empty vector. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated. Lane 1: normal Aα, Bβ, and γ; lane 2: normal Aα, normal Bβ plus mutant G444S Bβ, and normal γ; lane 3: normal Aα, mutant G444S Bβ (only), and normal γ. (B) Multiple alignment of the C-terminal region of fibrinogen Bβ chain. Sequences from human (P02675), mouse (XP_130960), rat (P14480), bovine (P02676), chicken (Q02020), Xenopus (Q91589), and lamprey (P02678) were obtained from the Swiss-Prot database (except the mouse sequence, which was obtained from the National Center for Biotechnology Information [NCBI]) and aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw/). Conserved amino acids are highlighted in gray; arrows indicate the positions of previously reported mutations: L383R, G430D, Y447G, W467X, W470X. The open arrow indicates the position of the mutated glycine residue (G444S).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/13/10.1182_blood-2003-06-2141/6/m_h82435339002.jpeg?Expires=1769470303&Signature=4akycCue2hIPmbKSGlS7C2s5vmSnkYqBWtXxzhTZTbQTnofFhA8K1SMrd6W6GNWTzZe-T-G9MQRH3eZOBdkPrBjs2SFjfQFZhK6lBtT8lZW~DdHmM4xb3i36hE1~VCJpsRQFUSUxxTLU8tneyy4vispTcxhHFzKvBhK3B8BFqKKN08g6Z-dbkH3DF~O2qe8s0g~hZ1YN4xs~L5GcpmGXvS-uiZFrJaTLRh9GShaiefMHCf402xyuIp4FZddWCp09uWD8ta9O7Heiae7iobr48oUuUvTQ8xkNZlCgsbRw3EWMXghlfb0IxPPEhhRR~BsnCsbkgiLmG~7HNo7opKPzgg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
