Abstract
Different forms of interleukin-15 (IL-15) have been identified and shown to elicit different transduction pathways whose impact on hematopoiesis is poorly understood. We demonstrated herein that hematopoietic CD34+ cells constitutively produced endogenous secreted IL-15 (ES-IL-15) that activated different transcription factors and controlled the expression of several functional proteins, depending on the progenitor source. Thus, nuclear factor-κ B (NF-κ B) was activated in bone marrow (BM) and cord blood (CB) progenitors, whereas signal transducer and activator of transcription 3 (STAT3) and STAT5 activation was restricted to peripheral granulocyte—colony-stimulating factor (G-CSF)—mobilized and BM progenitors, respectively. ES-IL-15 acts through autocrine/paracrine loops controlled by high-affinity receptors involving IL-15 receptor α (IL-15R α). Furthermore, ES-IL-15 was found to differentially control the expression of several functional molecules important for hematopoietic differentiation. Indeed, in BM precursors, neutralizing anti—IL-15 monoclonal antibody (mAb) inhibits the expression of the γ c chain and of the chemokine stromal derived factor-1 (SDF-1) but had no effect on vascular cell adhesion molecule 1 (VCAM-1) and β 1 integrin adhesion molecule expression. Conversely, in CB progenitors, anti—IL-15 mAb inhibited VCAM-1 and β 1 integrin expression without affecting γ c chain expression and, most important, up-regulated SDF-1 expression. In conclusion, unprimed human hematopoietic CD34+ cells secrete cell-unbound IL-15, which activates through autocrine/paracrine loop distinct signaling pathways, depending on the progenitor source, thereby influencing the expression of several molecules important in the control of hematopoiesis. (Blood. 2003;102:109-117)
Introduction
Interleukin-15 (IL-15) is a cytokine that shares with IL-2 the use of the IL-2 receptor β/γc (IL-2Rβ/γc) signal transduction apparatus and several associated biologic activities.1,2 Unlike IL-2, the IL-15 gene is expressed in several cell types, and several posttranscriptional mechanisms control its synthesis, secretion, or both.1,2 Two IL-15 transcripts, generated by alternative splicing, encode the IL-15 preproteins 48L-IL-15 and 21L-IL-15, which display different signal peptides of 48 and 21 amino acids (aa's), respectively.3,4 The 48L-IL-15 isoform is targeted to the secretory pathway, but it exhibits a low secretion potential,5,6 whereas the 21L-IL-15 isoform appears to be restricted to the cytoplasm and nucleus.5,7 Moreover, certain cells such as macrophages constitutively express a bioactive membrane-bound IL-15 form, whose relationship to the 2 isoforms of IL-15 preprotein is still unclear.8 Nishimura et al9 suggested differential roles for the 2 isoforms in vivo using transgenic (TG) mice expressing either 48L— or 21L-IL-15. Thus, the 48L-IL-15 TG mice exhibited enhanced host defenses against Salmonella infection, whereas the 21L-IL-15 TG mice were susceptible and demonstrated impaired production of endogenous IL-15 and interferon-γ (IFN-γ).
In addition to the IL-2Rβ/γc complex, IL-15 binds to a private chain (IL-15Rα) that is ubiquitous, is present in 8 splicing variants, and displays, alone, very high affinity for its ligand. It has been proposed that IL-15Rα may act as a decoy receptor for excess IL-15 or may bind other receptor components not yet characterized.10,11 Lastly, the existence of a distinct high-affinity binding receptor, IL-15RX (60-65 kDa)—functionally independent of the IL-2Rβ/γc complex—has been reported in mast cells.12
IL-15 interaction with its receptor complex leads to a series of signaling events that are similar to those elicited by IL-2. These include activation of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway. IL-2Rβ is associated with JAK1, whereas γc is associated with JAK3, resulting in STAT3 and STAT5 phosphorylation, respectively. The IL-2Rβγ complex appears also to activate src-related tyrosine kinases, Bcl-2, and the Ras/Raf/mitogen-activated protein kinase (MAPK) pathway that ultimately results in fos/jun activation.1,2 Recently, it was proposed that the IL-15Rα is directly involved in signal transduction by activating the transcription factor nuclear factor (NF)—κB13,14 and the alternative receptor system IL-15RX to induce JAK2/STAT5 and tyrosine kinase 2/STAT6 (TYK2/STAT6) phosphorylation.12,15
IL-15 has pleiotropic activity that ultimately results in immunoregulatory cross-talk between the innate and the adaptive immune systems. Actually, IL-15 is considered the major physiologic growth factor responsible for natural killer (NK) cell differentiation of hematopoietic CD34+ progenitors and for survival and cytolytic activity of mature NK cells.2 With respect to IL-15/CD34+ cell interplay, IL-15 acts on hematopoietic precursors of different origin and various degrees of primitivity,16-21 resulting in the differentiation into NK cells and dendritic cells (DCs) and the expansion of long-term culture-initiating cells (LTC-ICs). Although most of these results have been obtained by using recombinant IL-15 (rIL-15), the role of IL-15 production in hematopoietic progenitors remains to be documented. In this respect, we have recently reported that BM hematopoietic CD34+ progenitors produced endogenous, secreted, cell-unbound IL-15 (ES-IL-15) involved in autocrine/paracrine loops.22
In the present study, we analyzed IL-15 production in freshly isolated hematopoietic CD34+ progenitor cells originating from various sources, including bone marrow (BM), granulocyte—colony-stimulating factor (G-CSF)—mobilized peripheral blood (MPB), and cord blood (CB). Furthermore, we determined the activation patterns of transcription factors and of functional molecules—for example, stromal cell—derived factor 1 (SDF-1), vascular cell adhesion molecule 1 (VCAM-1), β1 integrins, and γc chain—reported to play an important role in the regulation of hematopoiesis and whose expression might depend on IL-15—mediated molecular mechanisms. Thus, the SDF-1 chemokine, produced in vivo primarily by BM stromal cells, is a key factor in retaining human hematopoietic stem cells in the bone marrow during postnatal hematopoiesis by mediating chemokinesis and chemotaxis.23 In addition, SDF-1 is also produced by unprimed CD34+ progenitors, and it regulates, through an autocrine/paracrine mechanism, primitive hematopoiesis by suppressing apoptosis and promoting G0/G1 transition in CD34+ cells.24 On the other hand, the optimal expression of VCAM-1 and β1 integrin is, in vivo, essential for the adhesion between hematopoietic stem cells and stromal cells, as suggested by the aberrant circulation of CD34+ cells on the down-regulation or inhibition of these interactions.25-27 Furthermore, the IL-2Rγc chain plays a regulatory role in the balance between lymphopoiesis and myelopoiesis, as suggested by the inhibitory effects on lymphocyte maturation and myelopoiesis dysregulation observed in mice and in humans bearing γc or JAK3 deletions or mutations.28-30 Our data allowed us to discriminate differences between transcription factor activation and these functional molecules, all of which are important for the development of normal hematopoietic progenitors.
Materials and methods
Antibodies
Names and manufacturers of the various antibodies used in this study are listed in Table 1.
Purification of CD34+ cells
Mononuclear cells from cord blood (CB) or adult G-CSF—mobilized peripheral blood (MPB) were obtained from different donors and isolated after centrifugation on Ficoll gradient (Lymphoprep Nycomed Pharma SA, Oslo, Norway). Selection of CD34+ cells was performed using immunomagnetic selection (Miltenyi, Tebu, France) on mononuclear cells labeled with an mAb specific for the QBEND10 epitope of the CD34 antigen, as previously described.22 Normal BM CD34+ cells were isolated and purified as described.22,31 All cell types were analyzed soon after their immunopurification and in the absence of exogenous cytokines.
Cell lines
TF1β (IL-15Rα/β/γc) is an IL-15—dependent human hematopoietic CD34+ cell line that displays a pro-erythroid differentiation potential (generous gift of P. Sondel, Department of Human Oncology, University of Wisconsin, Madison).
Analysis of IL-15 transcripts by reverse transcription—polymerase chain reaction
Extraction of total RNA from CD34+ cells, cDNA synthesis, and amplification reactions were performed as previously described.3,6 Oligonucleotide primers and conditions used for amplification of IL-15 transcripts and control actin were as previously described.3,6 Amplified products were analyzed after electrophoresis on ethidium bromide agarose gels.
Confocal microscopic analysis of IL-15 and IL-15R
For double staining of IL-15 and the endosomal compartment, immunoselected CD34+ cells were preincubated in serum-free Iscove modified Dulbecco medium (IMDM) and then treated with Alexa Fluor488-transferrin at 37°C for 15 minutes. Subsequently, cells were washed and permeabilized with ORTHOpermeafix (Ortho Diagnostic Systems, Raritan, NJ) for 45 minutes at room temperature. Cells were then stained with the polyclonal rabbit antihuman IL-15 antibody p15, incubated for 30 minutes with a biotinylated goat antirabbit antibody, and finally exposed for 30 minutes to streptavidin-Alexa Fluor594.
After cell fixation and permeabilization, double indirect immunofluorescence was also performed to analyze the colocalization of IL-15R components. First, cells were stained in green with anti—IL-15Rα mAb M160 or anti—IL-2Rβ mAb CF-1 and in red with the rat anti—IL-2Rγ mAb TUGh4, followed by Alexa Fluor488-GAM and Alexa Fluor594-GAR incubation at room temperature. Interactions between γc chain and JAK3 were studied by using the rat anti—IL-2Rγ mAb TUGh4 (green staining) and the rabbit immunoglobulin G (IgG) anti-JAK3 (Upstate Biotechnology, Euromedex, Lake Placid, NY), followed by goat antirat IgG—fluorescein isothiocyanate (GAR-FITC) (Southern Biotechnology Associates, Birmingham, AL) and Alexa Fluor594-GAR (goat antirabbit) incubation at room temperature. After staining, cells were washed with phosphate-buffered saline (PBS), centrifuged in a Cytospin 3 (Shandon, Pittsburgh, PA), and analyzed by laser scanning confocal microscopy using a TCS Confocal System (Leica, Wetzler, Germany).
Analysis of signal transduction in hematopoietic CD34+ progenitors by confocal microscopy
Once purified, CD34+ cells were immediately analyzed for the expression of activated transcription factors using confocal microscopy. Half the samples were pretreated with anti—IL-15 mAb for 1 hours. Subsequently, cells were permeabilized, and indirect immunofluorescence was performed with antibodies recognizing the different phosphorylated transcriptional factors STAT3, STAT5, and STAT6 and the NF-κB subunit p65. Samples were then washed and incubated with Alexa Fluor488-GAR antibody. Nuclei were stained using propidium iodide (red staining). To detect the induction of NF-κB nuclear localization, MPB CD34+ cells (1 × 106) were preincubated in the presence or absence of anti—IL-15 mAb and were stimulated with 10 ng/mL tumor necrosis factor-α (TNF-α) (Biosource Int, Clinisciences, Montrouge, France) for 30 minutes.
Western blots
Experiments were carried out as described.12-15 Briefly, all cultures were serum-starved to reduce basal phosphorylation. Subsequently, cells (1 × 106) were washed twice and suspended in lysis buffer containing 0.5% NP-40. Cell lysates were analyzed by sodium dodecyl sulfate—polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% or 12% polyacrylamide gels) and were transferred to polyvinylidene difluoride (PVDF) membranes (NEN Research Products, Boston, MA). Membranes were blocked with 5% bovine serum albumin (BSA), probed with antibodies, and revealed with peroxidaseconjugated antibodies. Bands were visualized by an enhanced chemiluminescence (ECL) system (Amersham, Buckinghamshire, United Kingdom).
Flow cytometry analysis
Unprimed BM and CB CD34+ cells were incubated overnight in the presence of anti—IL-15 neutralizing antibodies or the NF-κB inhibitor BAY 11-7082 (Calbiochem, LaJolla, CA), a specific IB kinase inhibitor that prevents the degradation of IB and specifically abrogates NF-B DNA binding. These cells were then labeled as previously described,17 and cell surface proteins were analyzed using FACScalibur (Becton Dickinson). Mean fluorescence intensity was calculated with CellQuest software (Becton Dickinson).
Results
IL-15 is expressed in human CD34+ cells from various sources
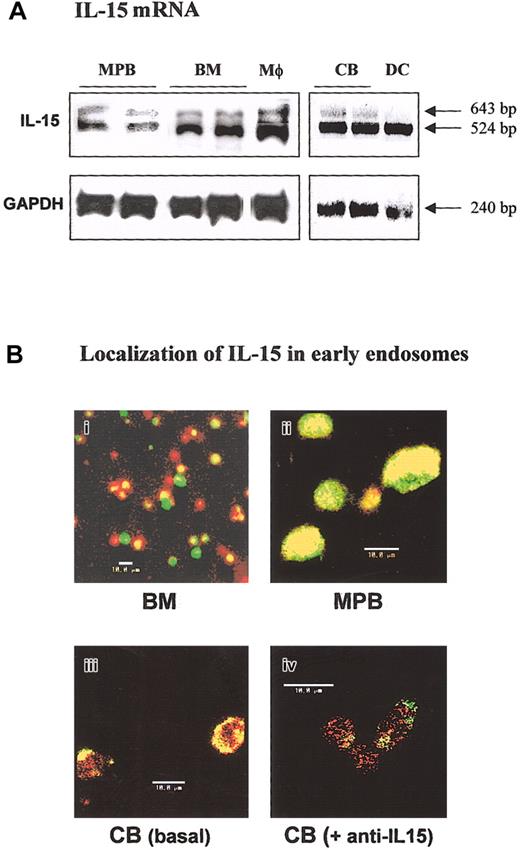
IL-15 transcripts were analyzed in purified BM, MPB, and CB CD34+ progenitors by RT-PCR. All cell types expressed both IL-15 transcripts generated by alternative splicing of exon A,3,4 even though the transcript encoding 48L-IL-15 was prominent (Figure 1A). As positive controls, lipopolysaccharide (LPS)—activated macrophages and DCs were used. Flow cytometric analysis indicated that CD34+ cells did not express membrane-bound IL-15 whatever their source (data not shown). Production of ES-IL-15 (red staining) and its localization in the early endosomes (green staining) were further confirmed in BM, MPB, and CB CD34+ cells by confocal analysis (Figure 1B1-3). The ES-IL-15 protein was clearly present at high levels in early endosomes, as shown by intense yellow staining, and its colocalization was totally inhibited by preincubation with anti—IL-15 blocking mAb (Figure 1B4), suggesting autocrine/paracrine ES-IL-15 trafficking. Nevertheless, the IL-15 amount in the supernatant was constantly below the enzyme-linked immunosorbent assay (ELISA) limit of detection and of biologic assays (3-10 pg/mL). As noted in previous observations in BM precursors,22 the 3 IL-15R subunits were to a large extent colocalized at the cell surface and inside the cells (data not shown).
Analysis of IL-15 expression in human CD34+ cells from various sources. (A) Expression of mRNA transcripts for IL-15 in human CD34+ cells by RT-PCR. Two amplification products for the IL-15 gene (643 and 524 bp) were detected in hematopoietic progenitors isolated from G-CSF—mobilized peripheral blood (MPB), bone marrow (BM), and cord blood (CB). As positive controls, LPS-activated macrophages (Mφ) and dendritic cells (DCs) were used. The housekeeping gene GAPDH (240 bp) was used as an internal control. (B) Confocal microscopic analysis of intracellular IL-15 localization in BM (i), MPB (ii), and CB (iii-iv) CD34+ cells by using anti—IL-15 mAb M111 (red staining) and FITC-transferrin as markers of early endosomes (green staining). The pictures are overlay-compacted images of serial optical sections of 1-μm thickness from the outside of the cells toward the inner compartments. Yellow staining at the submembrane level shows the localization of some endogenous IL-15 inside the early cell endosomes. The effects of neutralizing anti—IL-15 mAb on intracellular IL-15 localization in early endosomes are shown in lower panels iii and iv. The loss of yellow staining confirms that neutralizing mAb inhibits the retrapping of endogenously secreted IL-15 in CB CD34+ cells (panel iv). These data are representative of 3 different experiments.
Analysis of IL-15 expression in human CD34+ cells from various sources. (A) Expression of mRNA transcripts for IL-15 in human CD34+ cells by RT-PCR. Two amplification products for the IL-15 gene (643 and 524 bp) were detected in hematopoietic progenitors isolated from G-CSF—mobilized peripheral blood (MPB), bone marrow (BM), and cord blood (CB). As positive controls, LPS-activated macrophages (Mφ) and dendritic cells (DCs) were used. The housekeeping gene GAPDH (240 bp) was used as an internal control. (B) Confocal microscopic analysis of intracellular IL-15 localization in BM (i), MPB (ii), and CB (iii-iv) CD34+ cells by using anti—IL-15 mAb M111 (red staining) and FITC-transferrin as markers of early endosomes (green staining). The pictures are overlay-compacted images of serial optical sections of 1-μm thickness from the outside of the cells toward the inner compartments. Yellow staining at the submembrane level shows the localization of some endogenous IL-15 inside the early cell endosomes. The effects of neutralizing anti—IL-15 mAb on intracellular IL-15 localization in early endosomes are shown in lower panels iii and iv. The loss of yellow staining confirms that neutralizing mAb inhibits the retrapping of endogenously secreted IL-15 in CB CD34+ cells (panel iv). These data are representative of 3 different experiments.
ES-IL-15 controls STAT3, STAT5, and NF-κB activation in human CD34+ cells
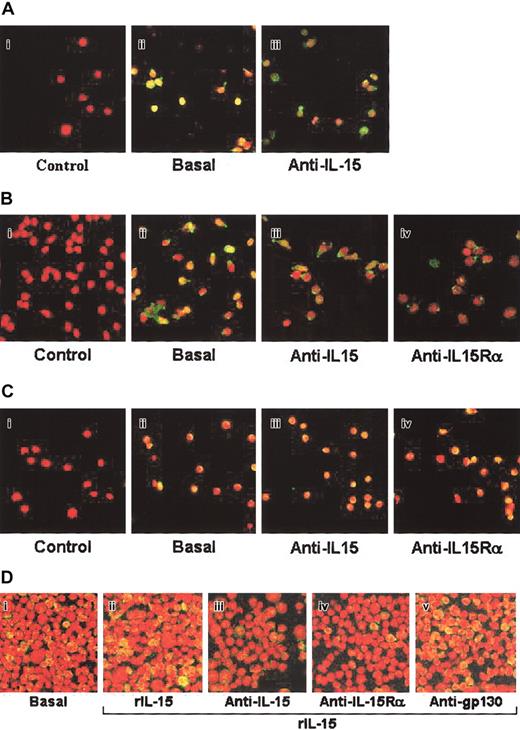
Western blot analysis showed that STAT3, STAT5, and the NF-κB inhibitor IκBα were constitutively phosphorylated in CD34+ cells from BM, MPB, and CB (Figure 2). By using blocking anti—IL-15 mAb, which efficiently blocked ES-IL-15 entry in the early endosomes (Figure 1B4), partial inhibition of STAT3 phosphorylation (-50%) was observed only in BM precursors. This treatment almost totally (-90%) inhibited STAT5 phosphorylation in BM and MPB precursors but not in CB CD34+ cells, whereas efficient inhibition of IκBα phosphorylation was observed in BM (-60%) and CB CD34+ cells (-73%) and less in MPB progenitors (-39%). These data suggest that in human CD34+ cells, ES-IL-15 controls the phosphorylation of different transcription factors depending on the progenitor's source. Subsequently, to identify the IL-15R subunits involved in the autocrine/paracrine loops, we treated BM- and CB-derived CD34+ cells with neutralizing mAbs recognizing the α and β subunits of the IL-15R. As isotype-matched control, we used neutralizing mAb directed against the gp130 subunit of the IL-6R (Figure 1D-E). Constitutive STAT3 phosphorylation was efficiently inhibited by anti—IL-15Rα and anti—IL-15Rβ mAbs (80% and 60%, respectively) in BM but not in CB CD34+ cells. Anti-gp130 mAb had no significant effect. These data strongly suggest that ES-IL-15 acts through autocrine/paracrine loops involving a high-affinity receptor. Indeed, the inhibition of STAT3 phosphorylation obtained using neutralizing anti—IL-15Rα alone indicates that the low amount of ES-IL-15 produced by hematopoietic progenitors must bind to the high-affinity heterotrimer IL-15Rα/β/γc to display biologic activity.
Analysis of transcription factor phosphorylation by Western blotting in human CD34+ cells from various sources. Hematopoietic progenitors isolated from BM (A), MPB (B), and CB (C) were incubated or not (basal) with neutralizing anti—IL-15 mAb for 1 hour. BM (D) and CB (E) CD34+ cells were also incubated with neutralizing anti—IL-15Rα, IL-15Rβ, or anti—IL-6R gp130 (isotype-matched control) mAbs for 1 hour. Cell extracts were analyzed by Western blotting using anti—phospho-STAT3 (pSTAT3), -STAT5 (pSTAT5), and -IκBα (pIκBα) antibodies (upper blots). Each membrane was reprobed with antibodies recognizing the native proteins or the β-actin (lower blots), showing equal amounts of protein in each sample. These data are representative of 3 different experiments.
Analysis of transcription factor phosphorylation by Western blotting in human CD34+ cells from various sources. Hematopoietic progenitors isolated from BM (A), MPB (B), and CB (C) were incubated or not (basal) with neutralizing anti—IL-15 mAb for 1 hour. BM (D) and CB (E) CD34+ cells were also incubated with neutralizing anti—IL-15Rα, IL-15Rβ, or anti—IL-6R gp130 (isotype-matched control) mAbs for 1 hour. Cell extracts were analyzed by Western blotting using anti—phospho-STAT3 (pSTAT3), -STAT5 (pSTAT5), and -IκBα (pIκBα) antibodies (upper blots). Each membrane was reprobed with antibodies recognizing the native proteins or the β-actin (lower blots), showing equal amounts of protein in each sample. These data are representative of 3 different experiments.
Because the phosphorylation of STAT factors is followed by their dimerization and nuclear migration, we asked whether anti—IL-15 mAb could also inhibit this additional activation step. The nuclear translocation of the transcription factor STAT3 was studied by using an antibody that specifically recognized the phosphorylated pSTAT3 but not the native protein.
Confocal microscopic analysis revealed that pSTAT3 was expressed in the cytoplasm (green staining) and at the nuclear level (yellow staining) in BM, MPB, and CB CD34+ cells (Figure 3). In BM and CB precursors, more than 90% of the cells showed nuclear localization of pSTAT3, whereas in MPB CD34+ cells approximately 70% of the cells were scored as positive. Blocking anti—IL-15 mAb inhibited pSTAT3 nuclear localization in BM and MPB (12% and 10% of residual positive cells, respectively), but not CB CD34+ cells, without affecting the intensity of the cytoplasmic staining. Identical inhibitory patterns were observed using neutralizing anti—IL-15Rα mAb. As a control of specificity, we used the IL-15—dependent CD34+ TF1β cell line, which displays a high-affinity IL-15R (IL-15Rα/β/γc) (Figure 3D). In these cells, recombinant IL-15 at 10 ng/mL induces the nuclear localization of pSTAT3, which is inhibited by neutralizing anti—IL-15 or anti—IL-15Rα mAbs, but not by an isotype-matched mAb recognizing the gp130 subunit of the IL-6R.
Analysis of phospho-STAT3 nuclear localization in human CD34+ cells from various sources by confocal microscopy. Hematopoietic progenitors isolated from BM (A), MPB (B), and CB (C) were incubated or not (basal) with neutralizing anti—IL-15 or anti—IL-15Rα mAbs for 1 hour and were analyzed for pSTAT3 expression and nuclear localization. Original magnification, × 63. The cells were permeabilized and labeled with anti-pSTAT3 antibodies, followed by incubation with Alexa Fluor488-GAR antibody. Propidium iodide stained nuclei in red, whereas phospho-STAT3 stained in green. As negative controls, cells were incubated with the second reagent and propidium iodide. Only nuclei were revealed (red staining). As a control for specificity, we used the IL-15—dependent CD34+ TF1β cell line (D), which displays high-affinity IL-15R (IL-15Rα/β/γc). These cells were treated for 1 hour with the neutralizing anti—IL-15, anti—IL-15Rα, or anti-gp130 subunit of the IL-6R (isotype-matched control) mAbs. Subsequently, they were incubated with recombinant IL-15 at 10 ng/mL and were analyzed for the nuclear localization of pSTAT3. STAT5 is not shown because anti—IL-15 mAb inhibits its phosphorylation. These data are representative of 3 different experiments.
Analysis of phospho-STAT3 nuclear localization in human CD34+ cells from various sources by confocal microscopy. Hematopoietic progenitors isolated from BM (A), MPB (B), and CB (C) were incubated or not (basal) with neutralizing anti—IL-15 or anti—IL-15Rα mAbs for 1 hour and were analyzed for pSTAT3 expression and nuclear localization. Original magnification, × 63. The cells were permeabilized and labeled with anti-pSTAT3 antibodies, followed by incubation with Alexa Fluor488-GAR antibody. Propidium iodide stained nuclei in red, whereas phospho-STAT3 stained in green. As negative controls, cells were incubated with the second reagent and propidium iodide. Only nuclei were revealed (red staining). As a control for specificity, we used the IL-15—dependent CD34+ TF1β cell line (D), which displays high-affinity IL-15R (IL-15Rα/β/γc). These cells were treated for 1 hour with the neutralizing anti—IL-15, anti—IL-15Rα, or anti-gp130 subunit of the IL-6R (isotype-matched control) mAbs. Subsequently, they were incubated with recombinant IL-15 at 10 ng/mL and were analyzed for the nuclear localization of pSTAT3. STAT5 is not shown because anti—IL-15 mAb inhibits its phosphorylation. These data are representative of 3 different experiments.
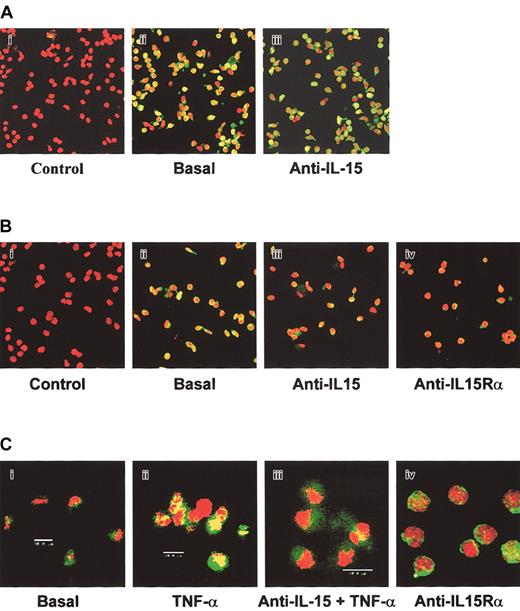
No pSTAT6 was detected in any of the progenitor types, indicating that it was not controlled by IL-15 (data not shown). Because IκBα phosphorylation is followed by the nuclear translocation of the NF-κB subunit RelA, we investigated its cytoplasmic expression (green staining) and nuclear localization (yellow staining). In BM and CB precursors, an intense yellow staining showed expression and RelA nuclear localization that was inhibited, with a different efficiency, by neutralizing anti—IL-15 mAb in the 2 types of CD34+ progenitors (Figure 4). Indeed, in CB precursors, 90% of the cells in the field displayed an intense nuclear yellow staining. Neutralizing anti—IL-15 mAb strongly inhibited the intensity of the staining and the number of positive cells (20%). In BM precursors, 50% of the cells in the field exhibited RelA nuclear localization. On anti—IL-15 treatment, we observed a 30% decrease in the number of yellow cells.
Analysis of NF-κB p65 nuclear localization in human CD34+ cells from various sources by confocal microscopy. Hematopoietic progenitors isolated from BM (A), CB (B), and MPB (C) were incubated or not (basal) with neutralizing anti—IL-15 or anti—IL-15Rα mAbs for 1 hour and were analyzed by confocal microscopy (original magnification, × 63), for NF-κB p65 expression and nuclear localization. MPB progenitors were also stimulated with TNF-α for 15 minutes. Cells were permeabilized and labeled with anti—NF-κB p65 antibodies, followed by incubation with Alexa Fluor488-GARa antibody. Propidium iodide stained the nuclei in red, and NF-κB p65 stained in green. As negative controls, cells were incubated with only the second reagent and propidium iodide. These data are representative of 3 different experiments.
Analysis of NF-κB p65 nuclear localization in human CD34+ cells from various sources by confocal microscopy. Hematopoietic progenitors isolated from BM (A), CB (B), and MPB (C) were incubated or not (basal) with neutralizing anti—IL-15 or anti—IL-15Rα mAbs for 1 hour and were analyzed by confocal microscopy (original magnification, × 63), for NF-κB p65 expression and nuclear localization. MPB progenitors were also stimulated with TNF-α for 15 minutes. Cells were permeabilized and labeled with anti—NF-κB p65 antibodies, followed by incubation with Alexa Fluor488-GARa antibody. Propidium iodide stained the nuclei in red, and NF-κB p65 stained in green. As negative controls, cells were incubated with only the second reagent and propidium iodide. These data are representative of 3 different experiments.
In contrast, no constitutive RelA nuclear localization was detected in MPB CD34+ cells. However, treatment with TNF-α strongly induced RelA nuclear localization, which was totally blocked by anti—IL-15 mAb. Identical inhibitory patterns were observed using neutralizing anti—IL-15Rα mAb. These data indicate that human hematopoietic precursors constitutively secrete ES-IL-15 that, through autocrine/paracrine loops controlled by high-affinity receptors, activates several transcription factors with patterns dependent on the source of CD34+ cells.
ES-IL-15 controls γc chain, VCAM-1, β1 integrin, and SDF-1 chemokine expression in human CD34+ cells
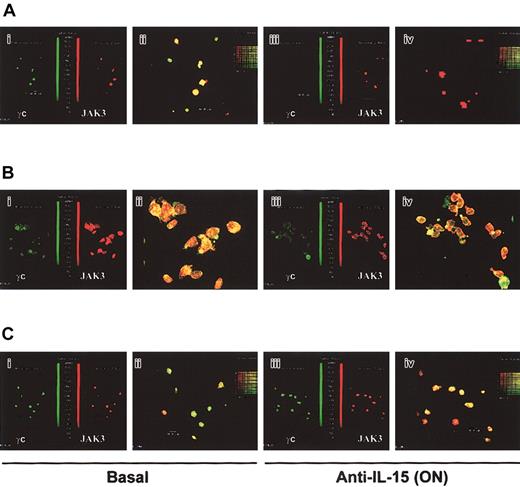
Expression of the γc chain (green staining) and interaction with its private kinase JAK3 (red staining) were analyzed in BM, MPB, and CB CD34+ cells by confocal microscopy. The 3 types of CD34+ cells constitutively expressed the 2 molecules, which appeared to be almost completely colocalized in BM and MPB cells but not in CB precursors, where the 2 molecules do not completely overlap (Figure 5). Moreover, their responses to neutralizing anti—IL-15 mAb differed. Thus, after 24-hour treatment with anti—IL-15—neutralizing mAb, inhibition of γc expression but not of JAK3 staining was detected in BM precursors, whereas no change in the expression of the 2 molecules was monitored in MPB or CB CD34+ cells. However, anti—IL-15 treatment prevented the colocalization of JAK3 and the γc chain in approximately 30% of MBP cells.
Analysis of the expression of the γc chain and JAK3 in normal human CD34+ cells and effects of anti—IL-15 mAb. Confocal microscopy on overlay pictures (original magnification, × 63) shows the presence of γc chain (green) and JAK3 (red) in BM, MPB, and CB CD34+ cells. Yellow staining demonstrates that the 2 molecules almost totally colocalize in BM (A) and MPB (B), but not in CB CD34+ cells, where there is only a partial overlapping of the 2 molecules (C). Side-side pictures of the previous picture shows that the expression of γc (totally) and JAK3 (partially) is inhibited after overnight treatment with neutralizing anti—IL-15 mAb in BM (Ai) but not in MBP (B1) and CB (C1) hematopoietic precursors. However, in MBP precursors, anti—IL-15 treatment prevents colocalization of the JAK3 and the γc chain in 30% of the cells in the field. These data are representative of 3 different experiments.
Analysis of the expression of the γc chain and JAK3 in normal human CD34+ cells and effects of anti—IL-15 mAb. Confocal microscopy on overlay pictures (original magnification, × 63) shows the presence of γc chain (green) and JAK3 (red) in BM, MPB, and CB CD34+ cells. Yellow staining demonstrates that the 2 molecules almost totally colocalize in BM (A) and MPB (B), but not in CB CD34+ cells, where there is only a partial overlapping of the 2 molecules (C). Side-side pictures of the previous picture shows that the expression of γc (totally) and JAK3 (partially) is inhibited after overnight treatment with neutralizing anti—IL-15 mAb in BM (Ai) but not in MBP (B1) and CB (C1) hematopoietic precursors. However, in MBP precursors, anti—IL-15 treatment prevents colocalization of the JAK3 and the γc chain in 30% of the cells in the field. These data are representative of 3 different experiments.
As illustrated in Figure 6, flow cytometry analysis showed that ES-IL-15 influenced the expression of molecules activated by the transcription factor NF-κB, which plays a major role in the control of hematopoiesis. In these experiments, BM and CB CD34+ cells were studied because ES-IL-15 was demonstrated to control the constitutive NF-κB activation only in these 2 types of progenitors (Figures 2 and 4). In BM progenitors, overnight treatment with neutralizing anti—IL-15 mAb totally abrogated constitutive SDF-1 production but did not modify VCAM-1 and β1 integrin expression. In contrast, neutralizing anti—IL-15 mAb up-regulated SDF-1 and totally inhibited VCAM-1 and β1 integrin expression in CB precursors. Of note, the specific NF-κB inhibitor BAY 11-7082 did not modify SDF-1 expression in CB cells.
Analysis of SDF-1, VCAM-1, and β1 integrin expression in normal human CD34+ cells and effects of anti—IL-15 mAb. Flow cytometry analysis shows the effect of overnight treatment with neutralizing anti—IL-15 mAb on the cytoplasmic expression of SDF-1, VCAM-1, and β1 integrin in BM (A) and CB (B) CD34+ cells. In CB progenitors, SDF-1 expression was also analyzed after cell treatment with the specific NF-κB inhibitor BAY 11-7082. Each figure is representative of at least 3 different experiments. Vertical bars indicate negative control intensity. Thin lines indicate basal expression, whereas solid bold lines indicate protein expression in treated cells.
Analysis of SDF-1, VCAM-1, and β1 integrin expression in normal human CD34+ cells and effects of anti—IL-15 mAb. Flow cytometry analysis shows the effect of overnight treatment with neutralizing anti—IL-15 mAb on the cytoplasmic expression of SDF-1, VCAM-1, and β1 integrin in BM (A) and CB (B) CD34+ cells. In CB progenitors, SDF-1 expression was also analyzed after cell treatment with the specific NF-κB inhibitor BAY 11-7082. Each figure is representative of at least 3 different experiments. Vertical bars indicate negative control intensity. Thin lines indicate basal expression, whereas solid bold lines indicate protein expression in treated cells.
Discussion
Normal unprimed hematopoietic progenitor cells secrete several hematopoietic factors that can regulate, through autocrine/paracrine loops, the development of normal hematopoietic cells.24,32-40 We show, herein, that freshly isolated human CD34+ hematopoietic progenitors purified from various sources, including BM, MPB, and CB, constitutively expressed the 2 IL-15 transcripts and produced very low amounts of a secreted, bioactive IL-15 form (ES-IL-15) that could play, in vivo, an important role in the early maturation of hematopoietic cells. Moreover, ES-IL-15 is shown to control, through high-affinity—receptor-dependent autocrine/paracrine loops, the activation of several transcription factors, including NF-κB, STAT3, and STAT5—but differently and according to the source of CD34+ cells.
This differential behavior could be associated with the differences in the nuclear localization of the intracytoplasmic IL-15 isoform or IL-15Rα that we have detected in the hematopoietic progenitors (data not shown). Indeed, studies on transgenic mice overexpressing the 2 IL-15 isoforms9 indicate that the intracellular IL-15 isoform, part of which is located in the nucleus,5,41 provides negative feedback on the production and functions of the secreted IL-15 isoform.9 Interestingly, the intracellular IL-15 isoform lacks a nuclear localization signal (NLS), whereas its private binding subunit—the IL-15Rα chain—possesses a potential NLS in its sushi domain, supporting potential direct action of the IL-15/IL-15Rα complex at the nuclear level.11 In this respect, we have recently shown that 2 human melanoma cell lines spontaneously expressing the 2 IL-15 isoforms display different intracellular trafficking, nuclear localization of the IL-15/IL-15Rα complex, and activation of the transcription factor NF-κB through this complex.41 The mechanism whereby intracellular IL-15 affects the production of the secreted isoform is unknown; however, taken together, these data suggest that the intracellular IL-15/IL-15Rα complex may affect transcription and functions of the secreted IL-15 isoform.9 Thus, it is possible that a different nuclear interplay between these molecules may differentially affect the behavior of hematopoietic precursors that equally express the 2 IL-15 isoforms and the 3 IL-15R subunits.
Through differential STAT5 activation, IL-15 might contribute to hematopoietic stem cell viability and proliferation that appeared to be decreased in STAT5-deficient mice.42 Alternatively, through the differential activation of NF-κB, which binds to specific sites in the promoter region of several factors regulating hematopoiesis,43 ES-IL-15 might control the production of β-chemokines and adhesion molecule expression. Indeed, our data show that ESIL-15 up-regulated SDF-1 expression in BM-derived hematopoietic progenitors, whereas an opposite effect was detected in CB CD34+ cells. However, in the latter cells, the use of specific NF-κB inhibitors did not alter SDF-1 expression, suggesting that endogenous IL-15 controls the production of the chemokine through mechanisms independent of NF-κB activation. Moreover, ESIL-15 neutralization totally inhibited the expression of the adhesion molecules VCAM-1 and β1 integrin in CB, but not in BM, hematopoietic CD34+ cells. This differential behavior is not surprising given that it has been reported that NF-κB may differentially control the same target in cells of different origin—such as TNF-α production in macrophages or B versus T lymphocytes.44,45 Finally, ES-IL-15 differentially controls γc chain expression. Indeed, by using neutralizing anti—IL-15 mAb, γc chain expression was inhibited in BM cells but not in CB and MPB CD34+ cells. In BM precursors, ES-IL-15 probably controls γc chain expression through posttranscriptional mechanisms, as reported for IL-2 in monocytes where this cytokine allows enhanced γc chain expression through mRNA stabilization.46
Even though recombinant and natural IL-15 produced by stromal cells induce NK cell differentiation of hematopoietic progenitors,6-22,47,48 ES-IL-15 is unable, by itself, to spontaneously induce this process. It may be that the complex interplay between different forms of IL-15 and CD34+ cells can influence the intercellular cross-talk that regulates hematopoiesis. Depending on their origin, normal precursors display different phenotypes, repopulating abilities after transplantation, and potentialities to generate functional NK cells of varied degrees of immaturity.49-53 All these properties are probably controlled by endogenously secreted factors acting through autocrine/paracrine loops.24,32-40 We propose that ES-IL-15, through the differential activation of several transcription factors and functional molecules, may influence some of these properties. For instance, SDF-1 up-regulation in BM precursors by ES-IL-15 could rely to the need for cell homing, whereas SDF-1 down-regulation in CB precursors could favor their status of circulating quiescent cells by limiting their G0/G1 transition. Moreover, because it has recently been shown that the CD34+ cell entrance in S-phase is prevented by the engagement of β1 integrins,54-56 we propose that the β1 integrin up-regulation in CB precursors by ES-IL-15 could provide an additional mechanism for cell maintenance in a quiescent state. Alternatively, ES-IL-15—induced up-regulation of adhesion molecules such as VCAM-1 and β1 integrins could explain the better adhesion of peripheral blood CD34+ cells to the stroma immediately after birth compared with their BM counterparts.57 Finally, because it has been suggested that γc may play a regulatory role in normal myelopoiesis independently of IL-2, IL-4, and IL-7,58 it is likely that ES-IL-15, through the differential regulation of this chain, could also control normal myelopoiesis. In this respect, it has been shown that BM hematopoietic progenitors express a subset of CD34+ cells γc chain-, specifically enriched in immature cells already committed to the erythroid lineage.59 In conclusion, ES-IL-15, through all these mechanisms, could differentially influence the specific regulation of hematopoiesis, depending on the anatomic localization and developmental stage of the hematopoietic progenitors.
Prepublished online as Blood First Edition Paper, March 6, 2003; DOI 10.1182/blood-2002-09-2760.
Supported by grants from the Association pour la Recherche sur le Cancer (ARC; grant 5801), NRB-Vaincre le Cancer, Groupement des Enterprises Françaises dans la lutte contre le cancer (FE.GE.FLUC), Fondation de France (99-003929), the Italian Association for Cancer Research, and the Italian Ministry of Health P.F. Biotechnology. J.G.-M. is the recipient of an ARC fellowship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.