The risk of hepatitis B virus (HBV) transmission by transfusion in sub-Saharan Africa is considered to be relatively low, and testing of blood donors is often not done or is done relatively poorly. To re-examine this attitude, we identified HBV chronically infected blood donors from a major hospital in Ghana with a range of hepatitis B surface antigen (HBsAg) assays. Test efficacy was estimated using HBV DNA as a gold standard, and the risk of HBV infection in blood recipients was estimated for different testing strategies. Particle agglutination, dipstick, and enzyme immunoassay (EIA) HBsAg screening detected 54%, 71%, and 97% of HBV infectious donors, respectively. The risk of HBV transmission to recipients less than 10 years old ranged between 1:11 and 1:326 with blood unscreened and screened by EIA, respectively. For older recipients, the risk decreased a further 4-fold because of the high frequency of natural exposure to HBV. A total of 98% of HBsAg-confirmed positive samples contained HBV DNA. HBV DNA load was less than 1 × 104 IU/mL in 75% of HBsAg-reactive samples, most of them anti-HBe reactive. Approximately 0.5% of HBsAg-negative but anti-HBc-positive samples contained HBV DNA. The use of sensitive HBsAg tests is critical to prevent transfusion transmission of HBV infection to young children in a population with a 15% prevalence of chronic HBV infection in blood donors. However, this will not have much effect on the prevalence of this infection unless other strategies to protect children from infection are also advanced in parallel.
Introduction
The prevalence of hepatitis B virus (HBV) chronic carriage in sub-Saharan Africa ranges between 3% and 22% in blood donors.1-3 Typically, more than 50% of blood donors and blood recipients have had natural exposure to HBV, and the need for hepatitis B surface antigen (HBsAg) screening of blood donations has often been considered of secondary importance because many donors are not infectious and many recipients are not susceptible.2At present, the World Health Organization (WHO) estimates that no more than 50% of the blood supply in sub-Saharan Africa is screened for HBsAg. This low rate of screening is due to lack of perceived utility, lack of funds, or both. No systematic study of donor and recipient populations has been undertaken that could provide the basic data to estimate the transfusion-related risk of HBV infection in high-prevalence areas of Africa.
To estimate the risk of transfusion-transmitted HBV infection in high-prevalence areas, we evaluated the relative efficacy of various HBsAg screening assays in blood donors from Kumasi, Ghana (an area with close to 15% prevalence of HBsAg).3 Sera from HBsAg-positive and -negative donors were tested for other serologic markers of HBV infection and for HBV DNA. To estimate the risk of HBV transmission by transfusion, sera from HBsAg-negative donors and from a group of potential blood recipients were tested for markers of existing infection.
Patients, materials, and methods
Donors
The blood center located at the Komfo Anokye Teaching Hospital in Kumasi, Ghana collects approximately 8000 units per year from 2 distinct populations of donors. Replacement donors are recruited from among the family or relatives of hospitalized patients who require a blood transfusion. Volunteer donors are recruited mostly from students aged 16 to 20 years and provide 15% to 50% of the blood supply, depending on the time of year. All donors were asked whether they had jaundice; a positive answer was cause for deferral. No consent form signature was requested from putative blood donors because this was not included in the Ghanaian Department of Health regulation for blood collection.
A predonation venous blood sample was collected from each donor with a 4-mL disposable syringe. Blood was transferred to a glass tube, allowed to clot, and centrifuged twice for 3 minutes at 1500 rpm with a table centrifuge. Before blood collection, potential donors were informed that HBsAg screening would be performed. During the session, volunteer donors identified as HBsAg reactive were personally notified that their blood could not be collected and were asked to attend the blood bank laboratory for confirmation of the test result. If the result was confirmed, the donor was offered a clinical examination, alanine aminotransferase (ALT) testing, and counseling.
Samples of 242 donations collected from 120 volunteer and 122 replacement donors over a one-week period were randomly selected for testing of additional HBV markers.
Patients
Whole-blood samples from 141 patients that had been collected for blood cell counts before blood transfusion were randomly selected from the hematology laboratory. The only additional selection criterion was an adequate sample volume. Sample volume tended to be lower in young children, who were therefore underrepresented. The samples were centrifuged, and supernatant plasma was collected and frozen at −20°C until tested.
HBsAg screening
Three distinct types of HBV screening assays were used during the study. During 2000, a latex agglutination assay was performed before donation according to the manufacturer's instructions (VEDAlab, Alençon, France). The sensitivity level provided by the manufacturer was 30 to 40 ng/mL HBsAg.
Between July and December 2001, every blood donor was tested for HBsAg before donation with a dipstick assay (VEDAlab) according to the manufacturer's instructions. The strip was dipped in the serum-containing glass tube for 10 seconds and read for the visibility of one test and one procedural control line after 10 minutes. The limit of sensitivity provided by the manufacturer was 5 ng/mL HBsAg.
The third type of assay used in the study was an enzyme immunoassay (HBsAg Murex EIA; Murex/Abbott, Dartford, United Kingdom). From October 1999 through March 2000, this assay was performed after donation according to the manufacturer's instructions. The sensitivity was less than 0.5 ng/mL. Initially reactive samples were retested, and the reactivity of both assays was taken as a positive result. Predonation samples reactive with the agglutination or dipstick assay were retested by EIA for confirmation. Samples nonreactive by rapid test but positive by EIA were retested in the Division of Transfusion Medicine virology laboratory in Cambridge with an alternative EIA (Bioelisa HBsAg; Biokit SA, Barcelona, Spain). The test sensitivity indicated by the manufacturer was 0.18 ng/mL HBsAg.
For some samples, Determine HBsAg (Abbott Laboratories, North Chicago, IL) was used.
Other HBV serologic markers
Additional HBV testing was performed on samples from blood donors and hospital patients. Anti-HBs testing was performed using the Murex/Abbott kit (Murex/Abbott, Dartford, United Kingdom) according to the manufacturer's instructions. An optical density corresponding to 10 mIU/mL or greater defined a positive result. When sample volume permitted, samples were also tested for hepatitis B e antigen (HBeAg), anti-HBe, and anti-hepatitis B core (HBc).
HBV DNA
Two sets of samples were tested for HBV DNA. The first set included 199 donor samples found to be positive for HBsAg by both agglutination and EIA. These were tested by transcription-mediated amplification (TMA; Gen-Probe, San Diego, CA [Dr C Giachetti]) and quantitative real-time polymerase chain reaction (QPCR Mx4000; Stratagene, San Diego, CA). The second set included 590 random donor samples that were negative for HBsAg by the agglutination assay. These were initially tested using the Gen-Probe TMA at Chiron (Emeryville, CA [Dr B Phelps and S Shyamala]). Positive samples were retested by QPCR.
HBV DNA detected by TMA
HBV DNA detection was accomplished by using research TMA reagents developed by Gen-Probe within the Chiron Procleix System. The TMA assay consists of 3 steps: target capture, target amplification, and detection of target amplicons following hybrid protection assay, all performed in a single tube. A total of 400 μL target capture reagent (TCR) (detergent solution buffered with HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] and containing capture oligonucleotides and magnetic beads) containing internal control and 500 μL of each specimen was pipetted manually into 10 tube units (TTUs). After addition of TCR and the test sample, the TTUs were incubated in a 60°C water bath for 20 minutes, followed by reverse-transcription incubation for 15 minutes. The rack of TTUs was then placed in the magnetic separator for 10 minutes, after which the liquid was aspirated from each tube and the beads were washed twice with 1.0 mL wash buffer to remove TCR, nonhybridizing components, and the rest of the specimen.
The captured target and the internal control were amplified by TMA. To the captured beads in each tube, we added 75 μL amplification reagent (primers, deoxyribonucleoside triphosphates [dNTPs], nucleoside triphosphates, and cofactors in Tris-buffered solution). Before the addition of 25 μL enzyme reagent (Moloney murine leukemia virus reverse transcriptase and T7 RNA polymerase in HEPES/Tris-buffered solution), the rack was incubated in a water bath at 60°C for 10 minutes, and then cooled at 42°C for 10 minutes. Immediately after addition of the enzyme reagent, the TTUs were shaken to mix and incubated in the water bath at 42°C for 60 minutes.
The RNA amplicons were detected by a hybridization protection assay with amplicon-specific acridinium ester-labeled probes. Following amplification, 100 μL probe reagent containing labeled oligonucleotides specific for HBV target and internal control was added to each tube, and tubes were incubated in a water bath at 60°C for 15 minutes. Following completion of the probe hybridization step, 250 μL alkaline selection reagent was added to each tube and then incubated at 60°C for 10 minutes to degrade the unhybridized probe. The tubes were then placed in a luminometer, which performs automatic injection of 200 μL auto detect 1 (0.1% hydrogen peroxide, 1 mM nitric acid) and 200 μL auto detect 2 (1 N NaOH). To determine reactivity, we measured the resulting chemiluminescence in relative light units (RLUs) and compared it with a cutoff value generated from the positive and negative calibrators included in each run using the Procleix system software. A sample with an analyte signal (RLU)/analyte cutoff (S/CO) ratio greater than 1 was considered reactive. An internal control contained in the TCR was added to each test specimen and assay calibrator reaction. The internal-control signal in each reaction was discriminated from the HBV signal by the differential kinetics of light emission. The 95% confidence limit of HBV DNA detection was estimated to vary from 50 to 15 IU/mL. The difference in sensitivity is due to the use of different lots of reagents with formulation adjustments during the development phase of the assay.
HBV DNA quantification
Viral DNA was prepared from 200-μL plasma samples using the QiaAmp blood and tissue kit (QiaGen, Crawley, United Kingdom) according to the manufacturer's instructions. HBV DNA was quantified using the Mx4000 Multiplex Quantitative PCR System (Stratagene). The sequences of the probe BS-1 and the primers HBV-Taq 1 and HBV-Taq 2 were designed from the conserved regions of HBV surface gene, as described previously.4 The fluorogenic probe was 5′-labeled with FAM (6-carboxyfluorescein) and 3′-labeled with TAMRA (6-carboxy-N-tetramethylrhodamine). Amplification was performed with the Brilliant Quantitative PCR Core Reagent kit (Stratagene). A QPCR reaction contained 1X core PCR buffer, 5 mM MgCl2, 0.8 mM each dNTP, 1.8 μM each primer, 0.2 μM fluorogenic probe, 3 × 10−4 mM reference dye (carboxy-X-rhodamine; ROX), 2.5 U SureStart Taq polymerase, and 10 μL template DNA preparation per 50-μL reaction. After an initial incubation at 95°C for 10 minutes, fifty 2-step cycles of 1 minute at 60°C and 30 seconds at 95°C were carried out. For each run, duplicates of a 10-fold serial dilution of WHO International Standard for HBV DNA for nucleic acid testing (NAT) assays 97/746 (National Institute for Biological Standards and Controls [NIBSC], Potters Bar, United Kingdom) containing 0.16 to 1600 IU HBV genome per reaction were analyzed. Based on the mean threshold cycles for each dilution, a linear regression was constructed with the threshold cycle as a function of the log of the amount of template molecules per reaction. Using this regression analysis, we calculated the number of template molecules per reaction based on the mean of duplicates for each plasma sample. The sensitivity of the method was estimated using the WHO standard 97/746 and probit analysis at 20 IU/mL.
HBV genome sequencing
Nested PCR was used to generate amplicons from the S gene using the primer combinations HBV-S1/HBV-S2 and HBV-S3/HBV-S4, as described previously.4 Conventional PCR was performed in 50 μL with 2.25 mM MgCl2, 0.2 mM each dNTP, 1 μM each primer, 1X PCR buffer II, 1 U AmpliTaq (Applied Biosystems, Warrington, United Kingdom), and 10 μL template DNA. Thirty-five cycles were performed; each cycle included denaturation at 95°C for 30 seconds, annealing at 48°C for 30 seconds, and extension at 72°C for 30 seconds. Sequences of amplicons were determined and phylogenetic analyses were performed using the Phylip software package (Phylogeny Inference Package version 3.5,http://evolution.genetics.washington.edu/phylip.html), as described previously.5
Statistical analysis
The probability that released donations would infect recipients was calculated for different donor sources, testing strategies, and recipient age groups (older and younger than 10 years) by multiplying the prevalence of HBsAg and/or HBV DNA positivity among donations by 1 minus the sensitivity of the test and then by the proportion of recipients expected to be susceptible to infection. The probable range around these point estimates was calculated using the upper and lower 95% confidence intervals of the proportions of donors with infections and the proportions of recipients who were susceptible (the test sensitivity was not varied).
Results
Predonation and postdonation screening for HBsAg
During the 6 months of the study conducted between 1999 and 2000, 2060 donations from replacement donors and 1527 from volunteer donors were tested after donation for HBsAg.3 The age of replacement donors ranged from 16 to 59 years (median, 32 years). The male-to-female ratio was 9.0. The age distribution was symmetrical around the median (33 for males and 27 for females). Volunteer donors ranged in age from 16 to 56 years (median, 18 years). The male-to-female ratio was 1.8. The age distribution was considerably skewed toward the younger ages; 65.5% of the population was younger than 20 years of age. During 2000, 7543 donors (45.5% volunteer) were screened before donation for HBsAg with the agglutination assay. In the second half of 2001, 3770 donors (54.1% volunteer) were screened before donation for HBsAg using the dipstick assay. In each donor category, the distributions of sex and age were similar to those of the population studied between 1999 and 2000.
Table 1 summarizes the results of HBsAg screening in the 3 blood-donor screening periods. The specificity of each assay was estimated on a subset of each population retested by Murex EIA for the reactive rapid tests and by an alternative EIA for the EIA-reactive samples. An increase in specificity was observed between agglutination and the other assays. The percentages of initially reactive samples confirmed were 85.6%, 95.1%, and 96.2% for agglutination, dipstick, and EIA, respectively. The confirmed HBsAg prevalence, adjusted according to the respective specificity of the assays, increased substantially with assay sensitivity, from 8.2% to 14.7% between the least sensitive agglutination assay and the most sensitive EIA.
To determine whether epidemiologic comparisons between the successive donor populations screened for HBsAg with different tests were legitimate, we studied volunteer and replacement donor ratio, sex ratio, and median age during the periods concerned. As shown in Table2, the donor populations were remarkably similar. The percentages of repeat donors were 7.5%, 7.4%, and 7.7% for 1999, 2000, and 2001, respectively.
Distribution of HBV serologic markers in blood donors and hospital patients
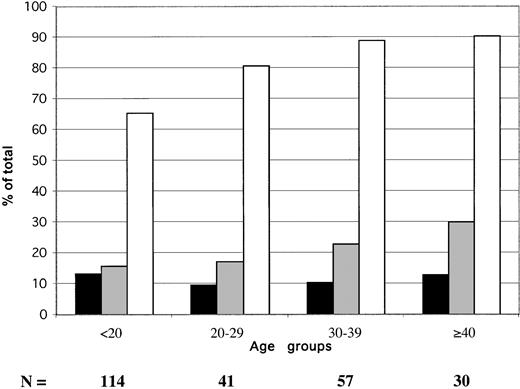
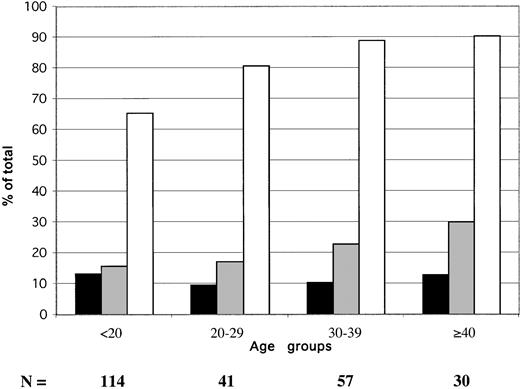
The age distribution of HBV serologic markers in 242 blood donors sampled in 2000 is shown in Figure 1. The prevalence of confirmed EIA-screened HBsAg did not vary significantly according to donor. It ranged between 9.6% and 13.2% (mean, 11.6%). The percentage of anti-HBs steadily, but not significantly, increased with age, from 16% to 30%. The prevalence of anti-HBc only (HBsAg and anti-HBs negative) samples (obtained by subtracting samples containing either HBsAg or anti-HBs from the total number of anti-HBc-positive samples) ranged between 35.1% in donors younger than 20 years and 48.4% in donors older than 40 years.
Age distribution of HBV serologic markers in 242 Ghanaian blood donors from Kumasi.
▪ indicate HBsAg; ░, anti-HBs; ■, anti-HBc. Anti-HBc includes HBsAg and anti-HBs-reactive samples.
Age distribution of HBV serologic markers in 242 Ghanaian blood donors from Kumasi.
▪ indicate HBsAg; ░, anti-HBs; ■, anti-HBc. Anti-HBc includes HBsAg and anti-HBs-reactive samples.
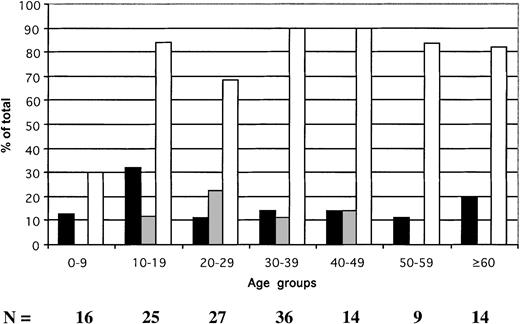
In hospital inpatients, the prevalence of HBsAg was 17.7% (25 of 141) (Figure 2), of anti-HBs was 10.5% (14 of 133), and of anti-HBc was 78.9% (86 of 109). The prevalence of HBsAg ranged between 11.1% and 32% according to age group. Anti-HBs was not found in patients younger than 10 years or in those older than 50 years and remained at low prevalence in other age groups. In contrast, the prevalence of HBV contact signaled by the presence of anti-HBc was lower in patients younger than 10 years than in the older age groups (P = .004). The prevalence of anti-HBc-only samples was 19% in patients younger than 10 and significantly higher in older patients (58.4%; P = .006). These data suggest that approximately 70% of the patients younger than 10 who were potential recipients of blood in the teaching hospital were susceptible to HBV infection. In contrast, 10% to 32% of older patients were at risk.
Age distribution of HBV serologic markers in 141 hospital patients identified as potential blood recipients.
▪ indicate HBsAg; ░, anti-HBs; ■, anti-HBc. The anti-HBc column includes samples reactive for HBsAg.
Age distribution of HBV serologic markers in 141 hospital patients identified as potential blood recipients.
▪ indicate HBsAg; ░, anti-HBs; ■, anti-HBc. The anti-HBc column includes samples reactive for HBsAg.
HBV DNA in HBsAg EIA-reactive Ghanaian blood donors
Before assessing the residual risk of HBV infection in seronegative donors, it was important to determine the correlation between HBsAg and other serologic markers and HBV DNA levels. This could provide clues for the interpretation of data.
HBV DNA and HBV serologic markers were studied in 199 samples found reactive for HBsAg by EIA in Ghanaian blood donors. The presence of HBsAg was not confirmed with an alternative EIA in 14 samples. However, 4 of these EIA-unconfirmed samples were found to contain low levels of HBV DNA ranging between 16 and 905 IU/mL. HBV DNA was first screened in confirmed HBsAg-containing samples with the qualitative TMA method on samples diluted 1:2 in phosphate-buffered saline, and 163 (88.1%) were found to be positive. Of these 163 positive samples, 140 were also tested by QPCR, and 138 (98.6%) were found to contain HBV DNA in concentrations ranging between 4 and 5.45 × 109 IU/mL. In addition, among the 22 TMA-negative samples, 16 of 19 tested by QPCR contained HBV DNA in concentrations ranging between 7 and 394 IU/mL (mean, 83.2; median, 49 IU/mL). Of a total of 159 HBsAg-confirmed positive samples tested by TMA and QPCR, 156 (98.1%) contained HBV DNA by at least one assay.
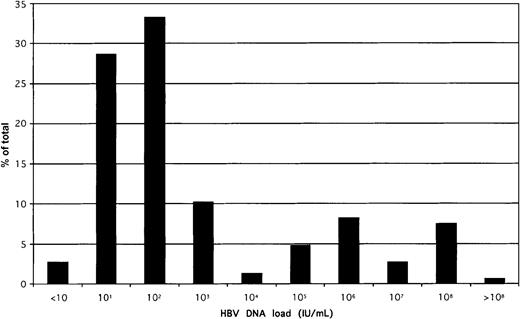
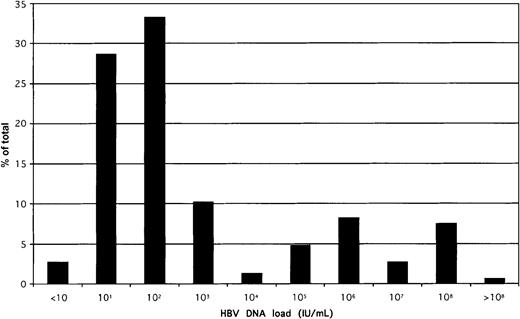
The distribution of HBV load in the whole population of 154 quantified samples is shown in Figure 3. A biphasic distribution was observed; that is, 75% of samples contained a viral load less than 10 000 IU/mL. Viral load was unrelated to sex (known in 142 donors) or to the ALT level (known in 89 donors). There was some decrease in viral load with increasing age; the trend, however, was not significant.
Distribution of HBV DNA load in 154 samples confirmed positive for HBsAg by EIAs.
The QPCR 95% confidence limit of detection was 20 IU/mL, but several samples could be quantified below that threshold. All samples were tested at least in duplicate, and mean values were taken for this analysis.
Distribution of HBV DNA load in 154 samples confirmed positive for HBsAg by EIAs.
The QPCR 95% confidence limit of detection was 20 IU/mL, but several samples could be quantified below that threshold. All samples were tested at least in duplicate, and mean values were taken for this analysis.
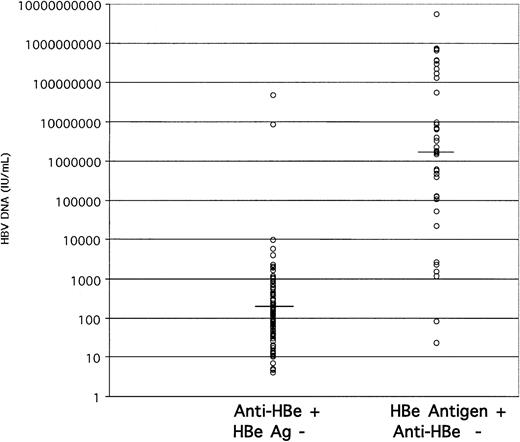
We next examined the correlation between viral load and the HBe antigen or antibody status of these donors. In 145 HBV DNA-containing samples of sufficient volume, HBe antigen only was present in 38 samples (26.2%), anti-HBe only in 100 samples (69%), both markers in 5 samples, and neither in 2 samples. The distribution of HBV load according to HBe status is shown in Figure4. In anti-HBe-positive samples, the viral load ranged between 4 and 9840 IU/mL, except for samples 72 and 74, which contained 8.6 × 106 and 4.83 × 107 IU/mL HBV DNA, respectively. In contrast, the HBV DNA load in HBeAg-reactive samples ranged between 23 and 5.5 × 109 IU/mL. The median viral load in the anti-HBe-reactive and -nonreactive groups was 1.4 × 102and 2.2 × 106 IU/mL, respectively. The 2 samples nonreactive for both markers contained 1.7 × 101 and 2.2 × 104 IU/mL HBV DNA.
Stratification of HBV DNA load according to HBe antigen marker status.
The median viral load for anti-HBe-carrying samples was 1.4 × 102 IU/mL and for HBeAg-carrying samples was 2.2 × 106 IU/mL (horizontal bars).
Stratification of HBV DNA load according to HBe antigen marker status.
The median viral load for anti-HBe-carrying samples was 1.4 × 102 IU/mL and for HBeAg-carrying samples was 2.2 × 106 IU/mL (horizontal bars).
Finally, we tested 67 HBV DNA-containing samples for the presence of anti-HBs. Six samples (9%) were reactive, with a viral load ranging between 17 and 143 IU/mL.
HBV markers in donations negative for HBsAg with the agglutination assay
To determine the residual risk of HBV transmission by transfusion after serologic screening with HBsAg assays of various levels of performance, we tested seronegative samples for HBV DNA. Blood donations that tested negative with the agglutination assay were retested with the Murex EIA. From a total of 590 samples, 14 (2.4%) were seropositive by EIA. Seven (1.2%) of these samples were positive for HBV DNA by TMA and therefore were not studied further. The 7 samples without detectable HBV DNA by TMA were tested by QPCR, and were all found negative. However, 3 of these 7 samples were reactive for anti-HBe, raising the possibility that a small number of HBsAg-containing samples did not contain detectable HBV DNA but corresponded to truly infected individuals or to false-positive results. The remaining 4 samples were considered EIA false positive.
Of the 576 seronegative samples, 37 were initially reactive by TMA. In these 37 samples, 15 samples had an S/CO less than 6.1 and 22 samples had an S/CO greater than 18 (range, 18.1-22.4). None of the 15 samples with low S/CO were positive by QPCR, and one sample was anti-HBe reactive. Of the 22 samples with an S/CO greater than 18, 12 samples contained HBV DNA quantifiable by QPCR. Eleven of these 12 samples had a low viral load (6-86 IU/mL), 3 of which were reactive for anti-HBe. The 12th sample reproducibly contained 4500 IU/mL HBV DNA and was negative for both HBe antigen and anti-HBe. One of the remaining 10 samples, which was positive by TMA but not by QPCR, was anti-HBe reactive.
When examining the correlation between HBV DNA load and anti-HBe, we found that 49 of 50 HBsAg-positive samples containing HBV DNA less than 100 IU/mL also contained anti-HBe (Figures 3-4). We therefore assumed that samples from donors at the tail end of HBV carriage (below the threshold of HBsAg detection), but containing HBV DNA detectable by both TMA and QPCR, were serologically positive for anti-HBe (and anti-HBc). In contrast, samples containing low levels of HBV DNA without serologic markers (anti-HBe and/or anti-HBc) were considered to be probable cross-contaminations that occurred during sample collection and preparation. The validity of this assumption was supported by the sequencing results in the S region of 3 samples that were processed at the same time in the blood bank. Sample 452 was HBsAg and anti-HBe reactive, and samples 454 and 455 were HBsAg and anti-HBe nonreactive. The HBV DNA loads were 7.3 × 106, 8.5 × 101, and 1.7 × 102 IU/mL, respectively. The sequences of the S region were identical.
In summary, among 590 samples from Ghanaian blood donations negative for HBsAg with the agglutination assay, 10 contained HBV DNA detectable by 2 assays and a serologic marker (7 HBsAg and 3 anti-HBe). Two more donations containing HBV DNA detectable by only one method and containing anti-HBe were considered of uncertain status. The 39 samples reactive serologically for HBsAg but without detectable HBV DNA (n = 7) or reactive for HBV DNA but without serologic markers (n = 32) were considered false positive or contaminated, respectively.
From these data, it was confidently concluded that 3 (0.5%) of 590 donations contained infectious HBV but remained undetected by HBsAg EIA. The 100% prevalence of HBV infectious donations in our donor population was therefore calculated as 14.7% (HBsAg positive) plus 0.5% containing HBV DNA as the only marker of active HBV infection, or 15.2%.
Relative sensitivity of screening assays for prevention of transfusion-transmitted HBV infection
From the results obtained in different sections of this study, the relative efficacy of the HBsAg or HBV DNA screening assays used can be extrapolated. Although the data for such extrapolations have been collected at different times between October 1999 and September 2001, the number of samples tested and the stability of the HBV epidemiology in the Kumasi area can be considered sufficient to make valid comparisons. It was considered that 100% of HBsAg-positive samples contained HBV DNA and were potentially infectious (Table 1) and that because of the large volume of whole blood transfused to patients (200 mL in pediatrics and 450 mL/unit in adults), detectable HBV DNA indicated infectivity by transfusion. To the prevalence of HBsAg in blood donations screened by EIA and confirmed (14.7%), 0.5% of HBV DNA found in HBsAg-negative donations was added, to reach 15.2% as the 100% prevalence of potentially infectious HBV blood donations. Of that total, EIA, dipstick rapid test, and latex agglutination assays were able to detect 96.6%, 71%, and 53.8%, respectively (Table1). These calculated test sensitivities, used along with the age-dependent susceptibility of putative blood recipients (Figure 2), resulted in estimates of the risk of HBV transmission by transfusion of blood, tested according to age and screening modalities, as shown in Table 3.
Discussion
Most developing countries are facing multiple threats to the safety of their blood supply. The cost of testing is one problem. If one excludes human immunodeficiency virus screening, which is essentially supported by long-term external aid, screening for hepatitis viruses can be done only if the cost of the tests is affordable by the patients and their families, who do not enjoy health coverage. The current assay technology is aimed at low prevalence in developed countries screening large numbers of samples with automated equipment. In circumstances in which a small percentage of collected blood is reactive with a screening assay, postdonation screening is a rational approach. In high-prevalence areas, up to 20% of the collected blood might be unsafe, testing errors or operational difficulties may have critical consequences,6 and the waste of blood and consumables is a heavy financial burden. It is clear that the guidelines for blood screening developed in and adopted by the affluent, low-prevalence countries are essentially unsuitable for resource-poor, high-prevalence countries. Improving blood safety necessitates conceptual changes, taking into consideration resources, assay technology, epidemiology, and blood-banking operations.
The results presented in this study indicate that the blood-donor population in Ghana includes approximately 15% chronic carriers of HBV, regardless of age, sex, or type of donor (Table 1), as previously reported.3,7 The high prevalence of HBV markers (anti-HBc) in donors (Figure 1) and in patients older than 10 years (Figure 2) is consistent with the concept that most primary HBV infections are transmitted either vertically or horizontally before age 10.8 9 The consistently low prevalence of anti-HBs contrasting with the high prevalence of anti-HBc in adults also suggests HBV infection occurring at an early age. A large proportion of infected individuals older than 10 no longer carry detectable anti-HBs, suggesting that most of those carrying anti-HBc as the sole marker of HBV infection are immune; the long interval between infection and testing accounts for the undetectability of anti-HBs. The apparent increase of anti-HBs prevalence with age observed in blood donors (Figure 1) might reflect re-exposure to HBV sexually or through other routes, including transfusion, in adults.
Two issues are critical: (1) How effective is HBsAg screening to remove infectious units? and (2) What proportion of the “anti-HBc”–only carriers remain infectious? The first issue was explored by comparing the efficacy of 3 types of HBsAg screening assays, the first 2 (agglutination and dipstick) being applicable to predonation screening. As shown in Table 1, considerable performance differences were observed, indicating that both latex agglutination and dipstick were insufficiently sensitive. Latex agglutination left nearly half of the presumably infectious units undetected. The prevalences of HBsAg reported in Table 1 are not directly comparable because each assay was used to screen blood donors during separate periods of time. Three elements, however, suggest that comparisons remain justified. First, the demographic characteristics of the populations of donors during the 3 time periods considered were remarkably similar in terms of type of donors (replacement or volunteer), age, and sex (Table 2). Second, the percentage of repeat donors in these populations was less than 8%, minimizing considerably a potential bias related to previously screened donors. Third, in a previous study,3 agglutination and EIA were tested in parallel (EIA was performed on agglutination-nonreactive samples), and EIA testing yielded more than 2% additional reactive samples. Although highly suggestive, the data shown in Table 1should therefore be considered indicative.
In further analyzing the implications of the differences in sensitivity observed among the screening assays, the quantitative data of HBV DNA in a subset of samples positive for HBsAg by agglutination and EIA were revealing (Figure 3). Approximately 75% of random chronic carriers had a low viral load (less than 10 000 IU/mL); most also carried anti-HBe (Figure 4). This explains why a relatively small difference in assay sensitivity (eg, 0.5-5 ng/mL between EIA and dipstick) translates into a 26% difference in the detection of infectious blood (Table 1). That 98% of HBsAg-containing samples also contain HBV DNA allows us to equate the presence of HBsAg and infectivity. This percentage of concordance is higher than reported by other investigators using QPCR10,11 or other quantitative methods,12who found no more than 84% DNA positivity in HBsAg-containing samples in asymptomatic carriers. The percentage of concordant results decreased to 77% or less in anti-HBe–positive samples. The difference is likely related to the higher sensitivity of our quantitative assay.
The data presented clearly indicate that latex agglutination and dipstick assays have insufficient sensitivity. However, they are both used in developing countries because of their low cost and flexibility to screen few samples without waste.
In low-prevalence populations such as in the United Kingdom or the United States, most blood donors carrying anti-HBc also carry detectable anti-HBs, and HBsAg-negative donations with anti-HBc as the only marker of HBV infection have been identified as an important source of posttransfusion HBV infection.13-15 In the Ghanaian population, with more than 40% of blood donations anti-HBc only, random HBsAg-negative blood donations were screened for the presence of HBV DNA. Interpretation of the results was made difficult because of the high probability that samples with high viral load may contaminate negative samples being processed nearby. We based our interpretation on the fact that a minute contamination would be detectable by PCR but not by serology, the dilution factor being sufficient to make anti-HBe or anti-HBc undetectable, but being overcome by the amplification capacity of PCR. This belief was supported by the results obtained from a group of 3 samples processed in the same batch. The sample containing a high viral load (7.3 × 106 IU/mL) was anti-HBc and anti-HBe positive, whereas the other 2 samples had low viral loads (85 and 167 IU/mL) and contained neither antibody. Identical S-region sequences found in all 3 samples strongly suggested cross-contamination. Considering the low likelihood of infection in the window period in the epidemiologic context of Ghana and the high probability of viral load greater than 103 IU/mL in such circumstances, samples containing lower levels of HBV DNA as the sole marker of the infection were considered cross-contaminations. Under these interpretation rules, 0.5% of EIA-seronegative samples contained both HBV DNA and antibodies to HBV.
In terms of blood safety, the issue is essentially the identification of infectious samples that would not be detected by HBsAg screening. According to the literature, this situation occurs in 3 separate types of situations: replacement mutations occurring in the domain that modifies antigenic recognition of the ‘a’ determinant16; deletions of various sizes in the pre-S1, pre-S2, or both regions17; and the masking of HBsAg detectability by immune complexes between the ‘a’ epitope and patient anti-HBs.18 Studies are in progress to determine which of these situations apply to some of our samples.
This study indicates that the detection of infectious HBV in blood donors in highly endemic areas such as Ghana is clearly useful for children, who represent 40% of transfusion recipients. Preventing infection by transfusion in these children is a worthwhile public health strategy only if they are not likely to acquire infection in childhood from other sources. The data presented suggest that the residual risk of posttransfusion infection resides essentially in chronic infections with low viral load and HBsAg level. To ensure blood safety, HBsAg testing requires highly sensitive assays enabling the identification of donors carrying low viral and antigen loads. Current EIAs, but not rapid tests, appear adequately sensitive.
We are indebted to Dr James Koziarz (Diagnostic Division, Abbott Laboratories, Abbott Park, IL), who supported part of the study. We are also indebted to Dr Julian Duncan (Murex/Abbott, Dartford, United Kingdom), who provided the anti-HBs and HBeAg assays. We are also indebted to Jillian Temple, Richard Larbi, Michael Adarkwa, and Yvonne Sekyie, who provided some technical help. Some of the HBV quantification kits by QPCR were gifts from Stratagene (San Diego, CA).
Prepublished online as Blood First Edition Paper, October 17, 2002; DOI 10.1182/blood-2002-04-1084.
Supported by grant BS01/01 from the National Blood Service, England.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jean-Pierre Allain, Division of Transfusion Medicine, Department of Haematology, East Anglia Blood Centre, Long Rd, Cambridge, CB2 2PT, United Kingdom; e-mail:jpa1000@cam.ac.uk.